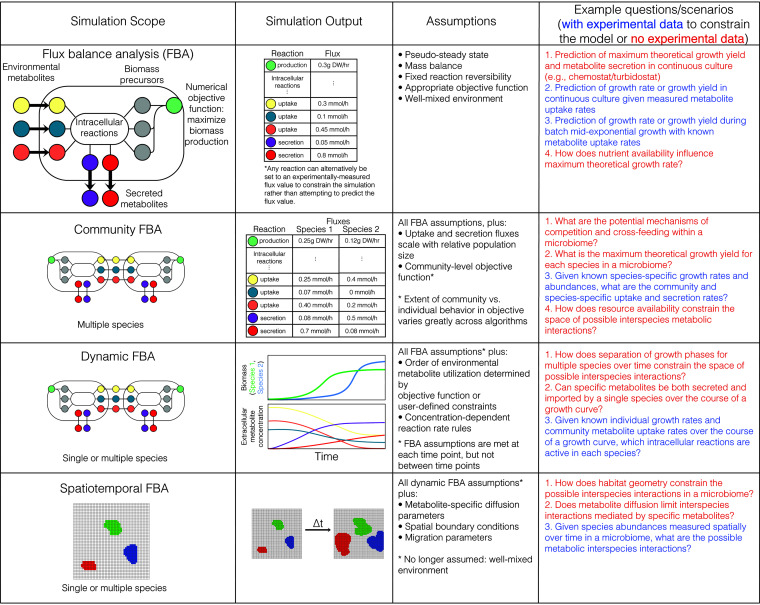
FIG 1.
Choice of microbiome modeling scope with genome-scale metabolic models (GEMs). Row 1 shows analysis of individual species (alone or within a microbiome) at a single time point with flux balance analysis (FBA). FBA can also be used for bag-of-genes models of a microbiota, which include all metabolic functions of the microbiota within a single compartment. Row 2 shows simulation of multiple species at a single time point in a community context with community FBA (where “Community FBA” is inclusive of all nondynamic FBA-based methods employing GEMs for multiple organisms in a shared extracellular compartment). Row 3 shows dynamic FBA, wherein metabolic fluxes and growth are simulated for a community with objective functions at the individual organism level. In dynamic FBA, multiple iterations of FBA are performed to introduce a temporal dimension. Community dynamics emerge in dynamic FBA through the shared extracellular compartment. Row four shows spatiotemporal FBA, which introduces spatial dimensions within a dynamic FBA framework to account for diffusion (or environmentally induced mixing) of metabolites and microbial biomass. Example simulation output and a nonexhaustive list of critical assumptions are listed for each scope. Examples of appropriate scenarios and questions are shown for each scope, with examples showing how introducing experimental data to constrain GEMs (text in blue) can increase the specificity of the simulation output at each scope relative to scenarios where little or no experimental data are used to constrain GEMs (text in red). Note that the following terms are not interchangeable: growth rate (growth per unit time), growth yield (growth per unit substrate uptake), and maximum theoretical growth yield (maximum growth per unit substrate, determined by stoichiometry and reversibility alone).