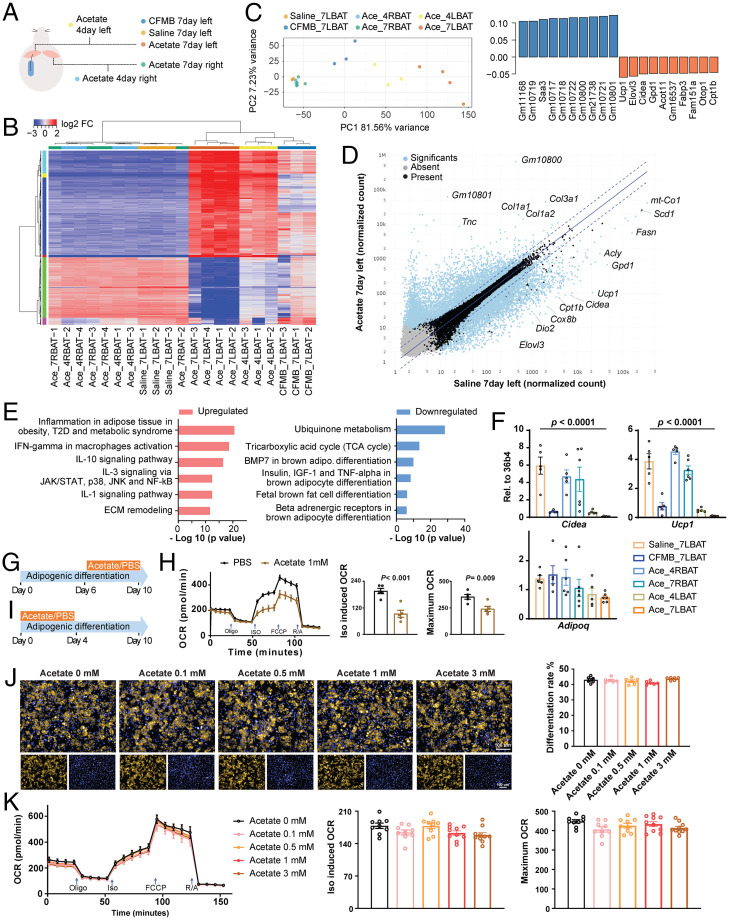
Fig. 2.
(A) Schematic illustration of the samples for RNA-seq analysis. Mice were implanted with catheter-guided pump delivering saline, acetate (30 mM), or CFMB (10 μM) to the left lobe of iBAT; the specific lobe of iBAT was collected after 4 or 7 d after implantation, as indicated. (B) Hierarchical clustering of RNA-seq data from iBAT samples is shown using a heatmap. The lengths of the branches represent the global differences between the samples, 2,000 genes with highest variance were included for the analysis, n = 3 to 4. (C) PCA of RNA-seq data (Left). Top 10 variable genes contributing to PC1 loading (Right). (D) Scatter plot of RNA-seq data, comparing acetate versus saline samples following 7 d of administration. Plotted are the normalized transcript counts. Significant differentially expressed genes (P < 0.01) are highlighted in blue, n = 3 to 4. (E) Metacore pathway analysis of up-regulated and down-regulated genes in iBAT samples from acetate versus saline administration, n = 3 to 4. (F) mRNA levels of Adipoq, Ucp1, and Cidea in iBAT samples analyzed by qRT-PCR, n = 5 to 6. (G) Schematic illustration for ex vivo adipocyte differentiation of iBAT stromal vascular fraction; acetate was supplemented from day 6 to day 10 of differentiation. (H) Time-resolved oxygen consumption rate in brown adipocytes treated with the acetate during differentiation days 6 to 10, n = 5. (I) Schematic illustration: acetate was supplemented from day 0 to day 4 of differentiation. (J and K) LD540 staining (J, n = 6) and time-resolved oxygen consumption rate (K, n = 9 to 10) in ex vivo differentiated cells from iBAT, treated with the indicated levels of acetate during differentiation from days 0 to 4.