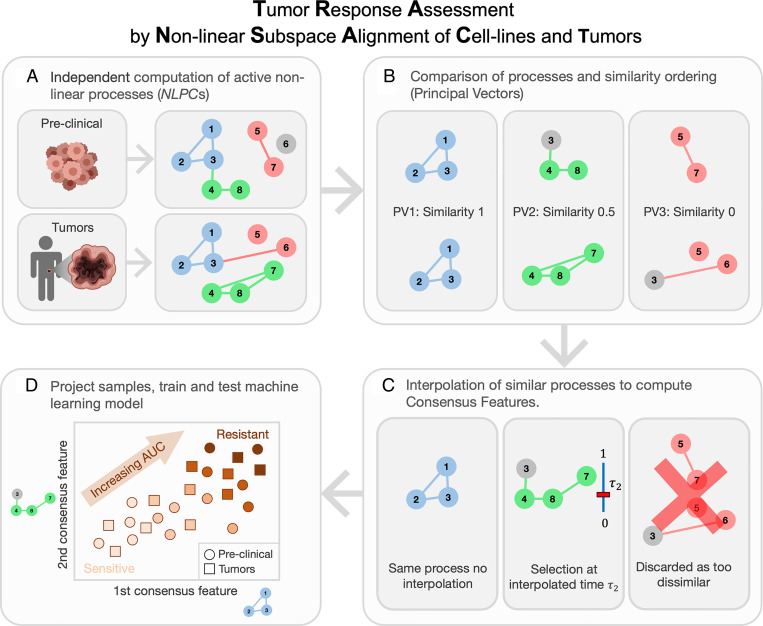
Fig. 1.
TRANSACT generates a nonlinear representation to transfer predictors of drug response from preclinical models to tumors. (A) In the first step, we use a nonlinear dimensionality reduction method to find biological processes active in preclinical models and in tumors. This step is performed independently in preclinical models and tumors and gives two sets of nonlinear processes called NLPCs. Here, we consider eight genes and three NLPCs for both preclinical models and tumors. A colored circle means that the corresponding gene contributes to the NLPC, while a gray circle is a gene that does not contribute. A colored connection is the interaction, or product, of two genes that contribute to the NLPC. For instance, the red preclinical NLPC represents the expression of Genes 5 and 7 and the product of the expression of Genes 5 and 7. (B) These two sets of processes are compared and ranked by similarity. For that purpose, we compute PVs, which are pairs of processes, one from preclinical, one from tumor, ordered by decreasing similarity. Here, the first PV is conserved between preclinical models and tumors, the second shows a 50% similarity, while the last PV corresponds to two distinct processes. (C) We first discard the PVs with low similarity, for example, PV 3. We then aggregate each PV pair into one consensus feature (CF) by finding an intermediate feature that balances the effect of the preclinical and tumor dataset. (D) Preclinical and tumor data are finally projected on each consensus feature, yielding a sample-by-CFs matrix. These CF scores represent the activity of biological processes essentially important for both preclinical models and tumors. These scores can then be used in any machine learning model to predict drug response.