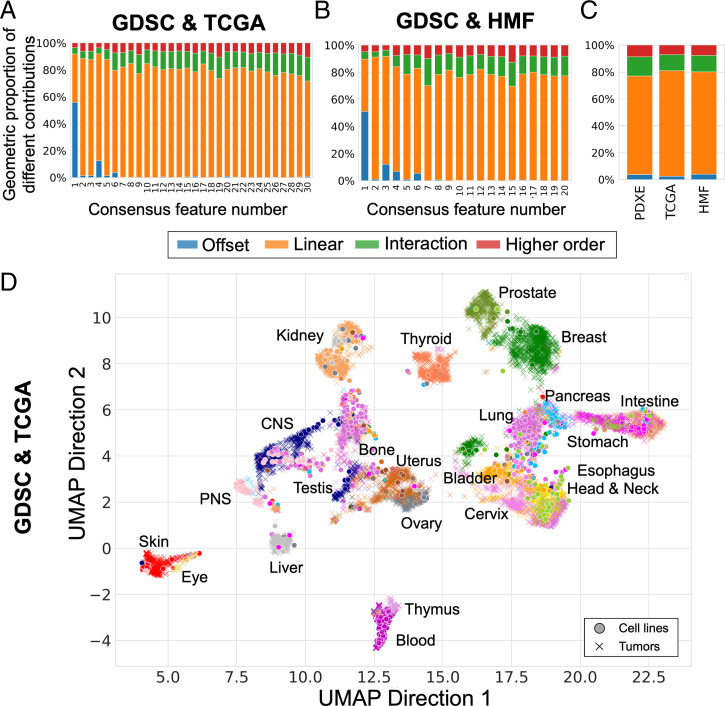
Fig. 3.
Pan-cancer consensus features between cell lines and tumors conserve tissue type information. We used cell lines (GDSC) as source data and compute two sets of consensus features with two different target datasets: primary tumors (TCGA, A and D) and metastatic lesions (HMF, B and E). (A) Proportion of linear and nonlinear contributions to each of the 30 GDSC-to-TCGA consensus features. (B) Proportion of linear and nonlinear contributions to each of the 20 GDSC-to-HMF consensus features. (C) Comparison of global contributions in the three analyses, that is, GDSC-to-PDXE, GDSC-to-TCGA, and GDSC-to-HMF. (D) UMAP plot of primary tumors (TCGA, 21 tissues) and cell lines (GDSC, 22 tissues) projected on the consensus features using the same parameters as selected in Fig. 2. The full legend for D and E is depicted in SI Appendix, Fig. 11.