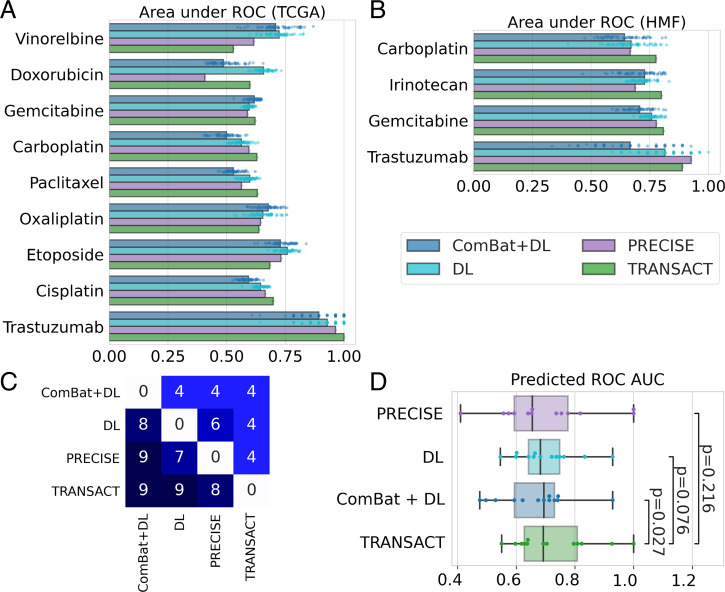
Fig. 4.
Consensus features improve response prediction in patients. (A) We used consensus features computed between the GDSC and each tumor dataset to train a predictor for 17 drugs on TCGA and six drugs on HMF using only GDSC drug response. We then predicted the continuous response in patients for each drug and compared this predicted value to the observed clinical response using a one-sided Mann–Whitney U test (Tables 1 and 2). We performed the same prediction tasks using three state-of-the-art approaches (ComBat+DL, DL, and PRECISE), and we summarized the results in a boxplot for TCGA (A) and HMF (B), restricting to the drugs in which at least one method reaches a significant prediction. For ComBat+DL and DL, we display the results obtained using 50 different random initializations. (C) Table of performance comparing each method; to read as “method on y-axis obtained higher AUC than method on x-axis,” for example, “PRECISE obtained higher AUC than ComBat+DL for nine drugs.” (D) Comparison of ROC AUCs obtained with each of the four methods for the 13 drugs with significant prediction. P values computed using a one-sided Wilcoxon paired rank-sum test.