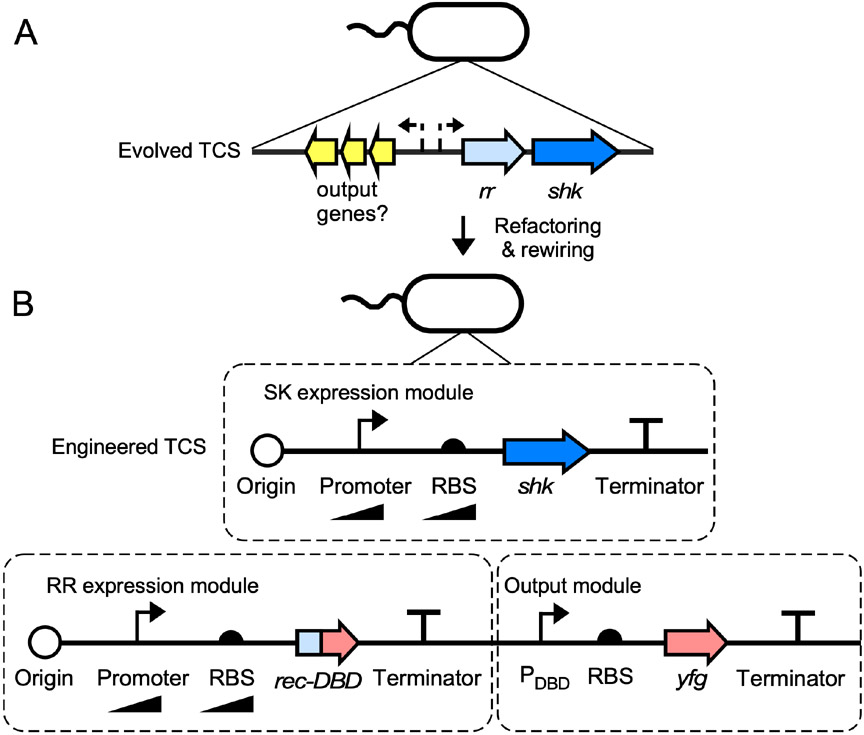
Figure 2.
Refactoring and rewiring TCSs. (A) Evolved TCSs can be computationally identified in bacterial genomes due to the conservation of SHK and RR domain architectures and the fact that the genes encoding interacting SHK-RR pairs often reside adjacent to one another. However, the promoters (dashed bent lines) and RBSs driving the expression of shk and rr genes can be difficult to predict and characterize from sequence information. This problem is exacerbated for output promoters, which may or may not reside adjacent to shk and rr genes. (B) TCSs can be engineered to function more reliably by replacing native promoters and RBSs with well-characterized synthetic versions. Promoters and RBSs of different strengths should be screened to achieve optimal SHK and RR expression levels. If an evolved output promoter is unknown or has undesirable features, it can be replaced by DBD swapping, wherein the native rr gene is replaced by a chimeric rec-DBD gene and the native output promoter is replaced by a well-characterized promoter that responds to the Rec-DBD protein (PDBD). yfg: your favorite gene.