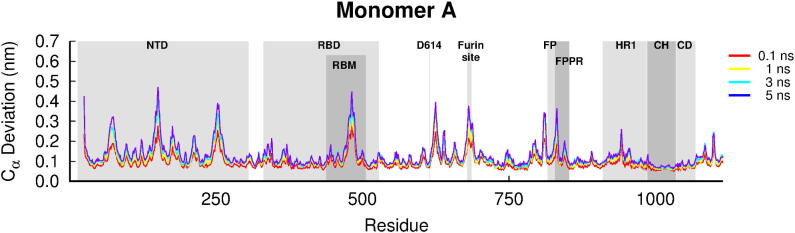
Fig. 2.
Average Cα-positional deviation for the first monomer in the five nanoseconds after LA removal from the FA sites in the SARS-CoV-2 spike. The structural deviations were calculated using the Kubo-Onsager approach [34], [35], [36], [37] for the pairwise comparison between the equilibrium LA-bound and dynamical-nonequilibrium apo spike simulations and averaged over all 90 replicates. A similar response to LA removal is observed for the other two monomers (Figure S4). Some relevant motifs are highlighted in grey, namely N-terminal domain (NTD), receptor-binding domain (RBD), receptor-binding motif (RBM), fusion peptide (FP), fusion-peptide proximal region (FPPR), heptad repeat 1 (HR1), central helix (CH) and connector domain (CD).