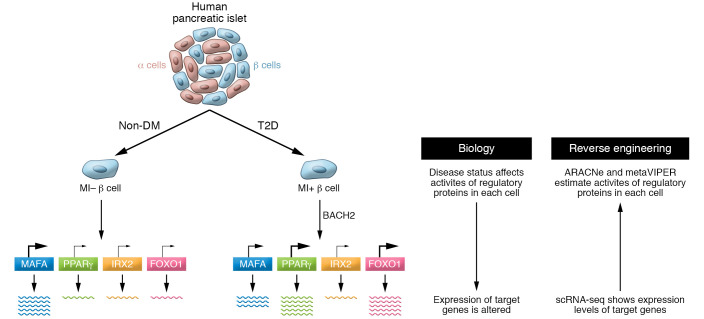
Figure 1. Reverse engineering of single-cell RNA sequencing data profiles regulatory protein activity in human islet cells.
The activity of regulatory proteins in each human islet cell changes, depending on factors such as cell identity (α vs. β cells) and disease status (nondiabetic [non-DM] vs. T2D). Son, Ding, et al. (7) used reverse engineering to extrapolate the activity of each regulatory protein from the expression level of target genes. Gene expression profiles were obtained by scRNA-Seq analysis, and regulatory protein activity was predicted using two algorithms, ARACNe and metaVIPER. Metabolically inflexible (MI) β cells showed transcription factor (TF) and co-TF activity changes, as exemplified in this model by increases in PPARγ and FOXO1 activities in T2D islets. Notably, BACH2 was implicated as one driver of T2D regulatory protein activities.