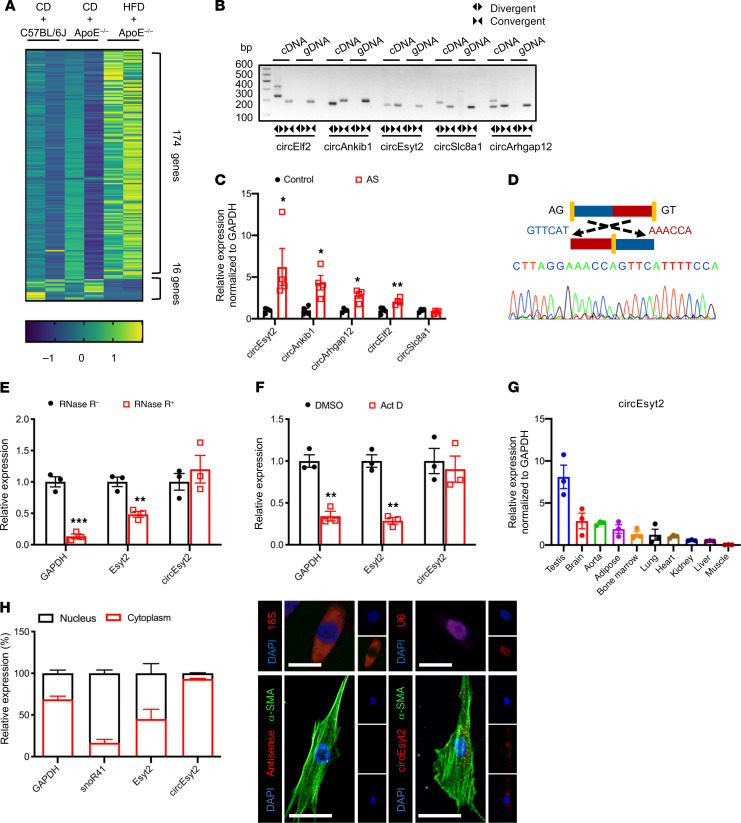
Figure 1. Identification of circEsyt2 based on circRNA profiling of mouse atherosclerotic aortae.
(A) Heat map of differentially expressed circRNAs in atherosclerotic (HFD + ApoE–/–) and control groups (CD + ApoE–/– or CD + C57BL/6J). CD: chow diet. HFD: high fat diet. ApoE–/–: ApoE knockout mice. n = 2. (B) PCR amplification using divergent or convergent primers against complementary DNA (cDNA) or genomic DNA (gDNA) from aortae. (C) qRT-PCR to check for the expression of circRNAs in aortae. *P < 0.05, **P < 0.01 vs. control. n = 4. (D) Confirmation of the backsplicing junction site within the circEsyt2 by Sanger sequencing. (E) mRNA expressions after RNase-R digestion in mouse VSMCs followed by qRT-PCR. **P < 0.01, ***P < 0.001 vs. RNase-R. n = 3. (F) Actinomycin D (Act D) treatment of VSMCs followed by qRT-PCR. Linear mRNA controls: GAPDH and Esyt2. **P < 0.01 vs. DMSO. n = 3. (G) qRT-PCR to check for the expression of circEsyt2 in mouse tissues. n = 3. (H) Subcellular localization of circEsyt2 in VSMCs. Left: qRT-PCR to check for the expression of circEsyt2, Esyt2, linear mRNA in the cytoplasmic and nuclear fractions; GAPDH, cytoplasmic control; snoR41, nuclear control. Right: images of FISH for circEsyt2 (red) and cofluorescence with α-SMA (green) and DAPI (blue). 18S, cytoplasmic control; U6, nuclear control; antisense, negative control. Scale bars: 20 μm. Data are mean ± SEM. Two-sided unpaired t test or Kolmogorov-Smirnov test for C, E, and F.