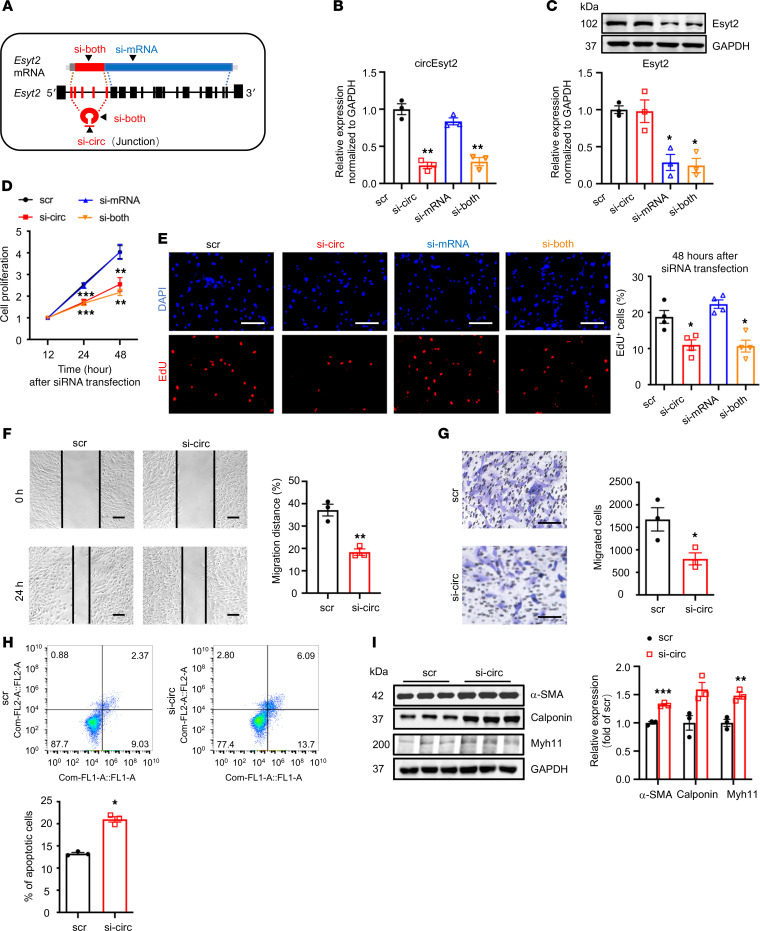
Figure 5. Effect of circEsyt2 silencing in HASMCs.
(A) Schematic representation of siRNAs designed for targeting circEsyt2 (si-circ), Esyt2 (si-mRNA), or both (si-both). Specifically, si-circ targets the junction formed by back-splicing of exons 6 and 2. Si-mRNA targets exon 7 (human) or exon 17 (mouse). Si-both targets exon 6 (human) or exon 4 (mouse). ex, exon. (B and C) Efficacy of circEsyt2 (si-circ), Esyt2 (si-mRNA) siRNA, or both (si-both) detected by qRT-PCR and Western blotting in HASMCs. Scrambled siRNA (scr) served as control. GAPDH, protein control. *P < 0.05, **P < 0.01 vs. scr. n = 3. (D) Cell counting kit-8 (CCK-8) assay of HASMCs treated with siRNAs, as shown in A for the indicated hours. **P < 0.01, ***P < 0.001 vs. scr. n = 4. (E) EdU incorporation assay of HASMCs, treated as in A for 48 hours. Left: representative immunofluorescence of EdU (red) and DAPI (blue). Scale bars: 100 μm. Right: percentages of EdU-incorporated HASMCs. *P < 0.05 vs. scr. n = 3. (F and G) Migratory ability assessed by wound healing (F) and Transwell assay (G) of circEsyt2-silenced HASMCs. Scale bars: 100 μm. *P < 0.05, **P < 0.01 vs. scr. n = 3. (H) Apoptosis detected by Annexin V–conjugated flow cytometry in circEsyt2-silenced HASMCs. *P < 0.05 vs. scr. n = 3. (I) Western blotting to check for the expression of α-SMA, Calponin, and Myh11 in circEsyt2-silenced HASMCs. *P < 0.05, **P < 0.01, ***P < 0.001 vs. scr. n = 3. Data are mean ± SEM. Two-sided unpaired t test for F–I. Two-way repeated measures ANOVA with LSD’s post hoc test for D. One-way ANOVA test with Dunnett’s T3 post hoc test for B, C, and E.