Fig 4.
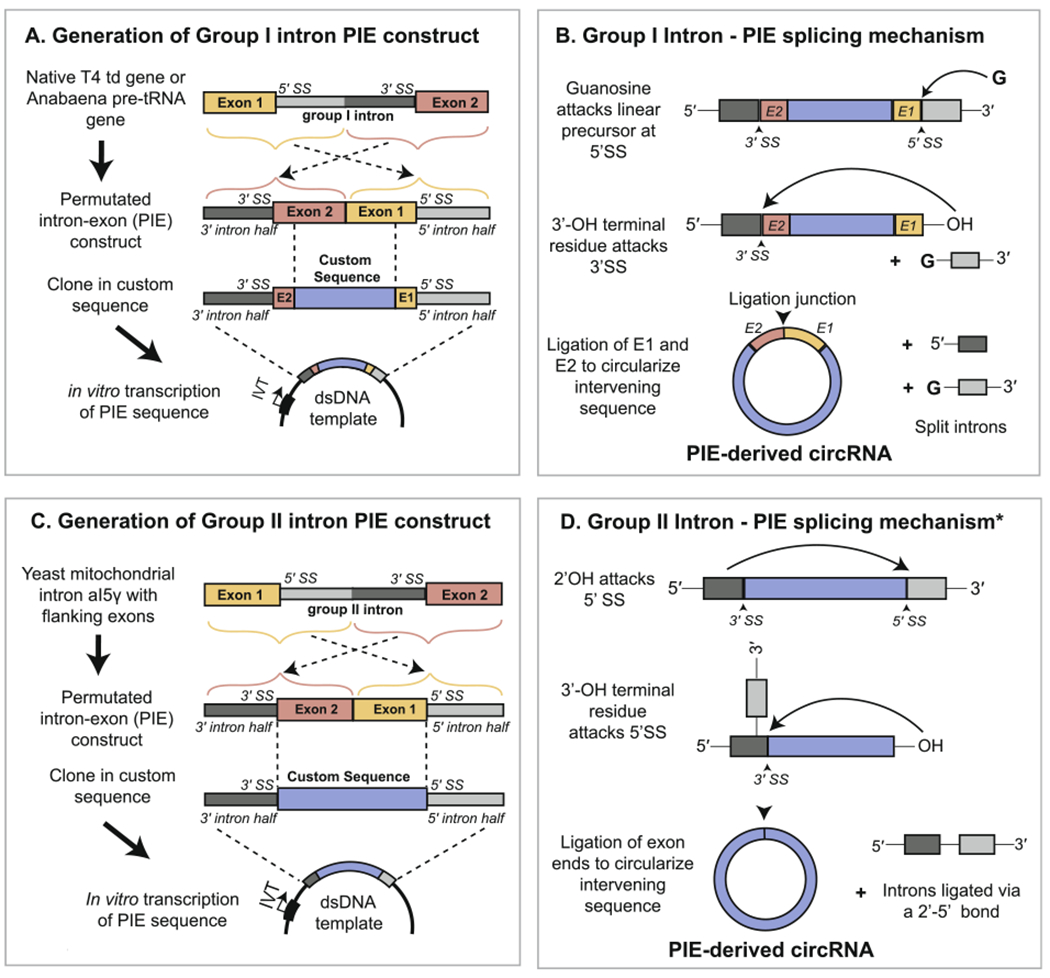
Generation of circRNA in vitro by the PIE method. (A) Generation of a PIE RNA construct from permutation of a native group I intron and insertion of a custom exonic sequence. (B) Upon in vitro transcription of the PIE sequence, circularization of the exonic region occurs auto-catalytically in the presence of free guanosine. A small portion of the flanking exonic sequence from the native group I intron-containing gene is retained at the ligation junction, which is formed from a 3′–5′ phosphodiester bond. (C) Generation of a PIE RNA construct from permutation of a native group II intron and insertion of a custom exonic sequence. Use of a Group II intron allows for the complete substitution of the native exonic sequence. (D) Upon in vitro transcription, circularization of the exonic region occurs auto-catalytically following release of the 5′ intron half (light gray). The asterisk signifies that the depicted mechanism, involving attack of the 2′ OH from the 5′ splice site, is one of multiple mechanisms by which exon release can occur. The resulting circRNA is ligated with a 3′-5′ bond and lacks remnants of the native group II intron-containing gene.
