Fig 8.
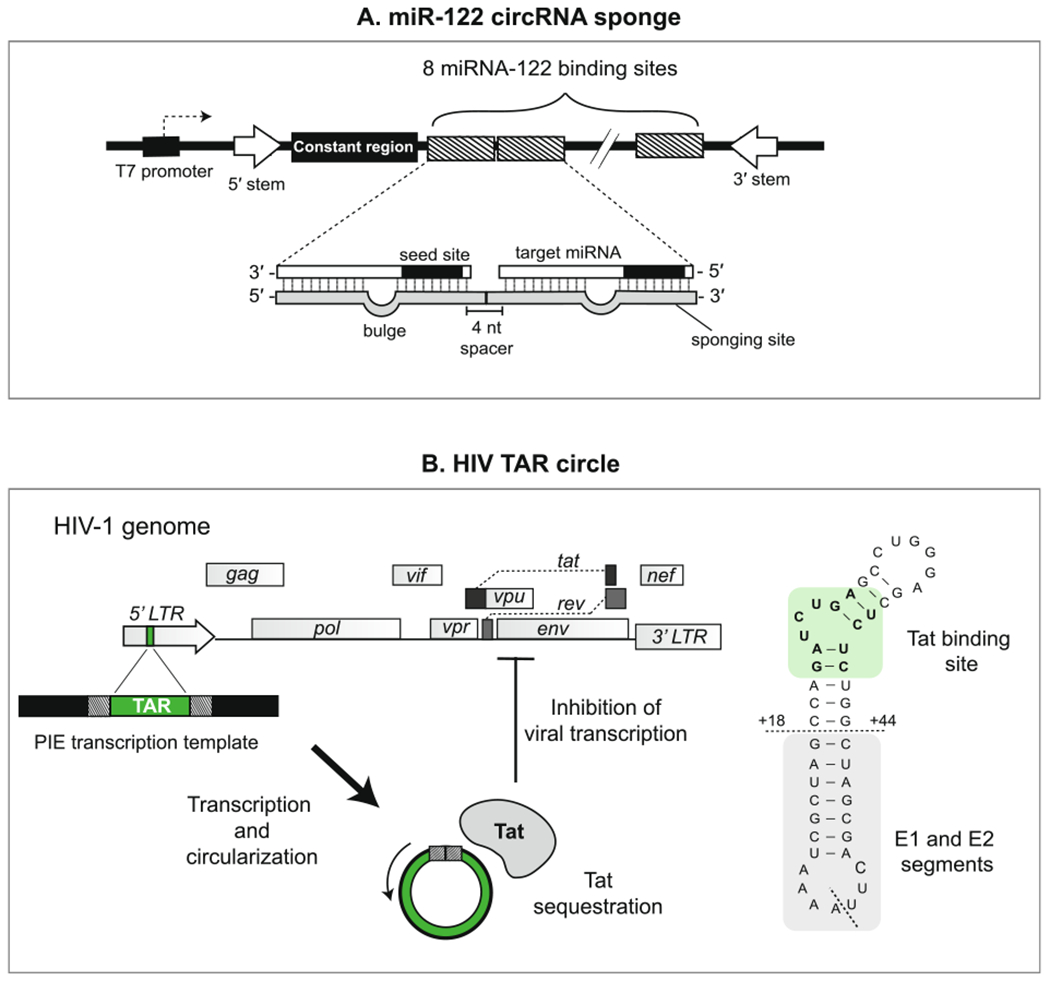
Design examples of miRNA- and protein-binding circRNAs. (A) Design of the minigene used to create a circular miR-122 sponge (ciRS-122) by Jost et al. (2018) [61,130]. 8 miR-122 sponge sites were concatenated, each exhibiting imperfect binding to miR-122 through introduction of a “bulged” region in the middle nucleotides. A constant region was included in the circularized sequence to facilitate detection of the circRNA sponge. Base-pairing sequences in the introns flanking the circularized region promote a stem-like secondary structure that facilitates circularization in vitro. (B) Design of the earliest protein-binding synthetic circRNA by Puttaraju and Been et al. (1992) [65,68,70]. The TAR sequence, found within the LTR promoter region of the HIV-1 genome, was incorporated into a PIE template to generate a synthetic RNA circle capable of binding the viral protein Tat. The sequence of the TAR circle (left) includes the Tat binding site (highlighted) and exonic sequences from the native group I intron (below horizontal dashed line). The ligation junction is indicated with a diagonal dashed line.
