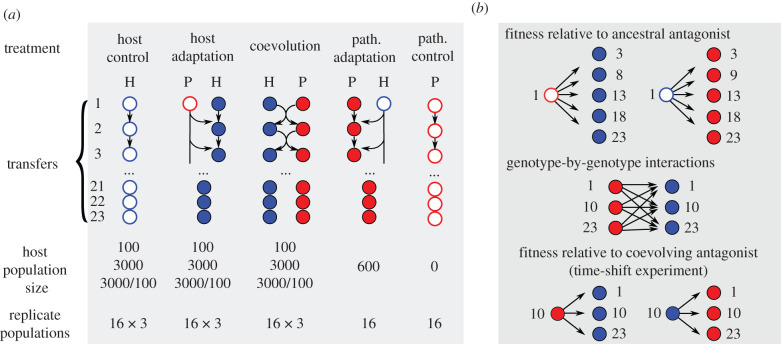
Figure 1.
Design of the evolution experiment and subsequent characterizations. (a) The evolution experiment consisted of five main treatments and three population size manipulations. Host and pathogen were evolved either in the presence of a coevolving antagonist (middle treatment), a non-adapting antagonist taken from a stock culture (second and fourth treatments), or in the absence of an antagonist (far left and far right treatments). The treatments with evolving hosts were repeated at a host population size of 100 (small populations), 3000 (large populations) or 3000 with periodic bottlenecks of 100 individuals at every fifth transfer (bottlenecked populations). We included 16 biological replicates for all treatment combinations. (b) Overview of subsequent characterizations. Evolved hosts and pathogens from transfers 3, 8, 9, 13, 18 and 23 were exposed to the respective ancestral antagonist (top illustration), allowing a comparison across the five main evolution treatments. We further performed time-shift experiments for the coevolution treatment, where hosts and pathogens from transfers 1, 10 and 23 were exposed to the co-adapting antagonist in all possible combinations (middle panel) or with a specific focus on the coevolved antagonist from transfer 10, in order to reconstruct the dynamics of coevolution. (Online version in colour.)