Table 1.
Examples of computationally derived molecules from drug repurposing for SARS-CoV-2.
| Structure | Drug | Method | In vitro/ in vivo activity | Reference |
|---|---|---|---|---|
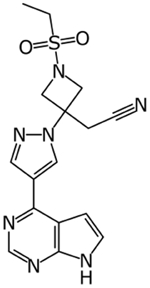
|
Baricitinib | knowledge graph | Significantly reduced viral load in human liver spheroids at 400 and 800 nM | [13–16] |
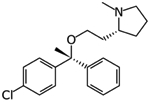
|
Clemastine | Protein interaction map | pIC50 5.67 | [23] |
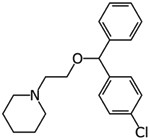
|
Cloperastine | Protein interaction map | pIC50 5.27 | [23] |
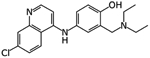
|
Amodiaquine | Ligand-based virtual screen | EC50 0.13 μM and 5.4 μM | [27] |
|
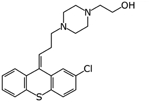
Zuclopenthixol | Ligand-based virtual screen | 1.35 μM and 15 μM | [27] |
|
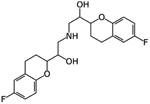
Nebivolol | Ligand-based virtual screen | 2.72 μM and 2.8 μM | [27] |
|
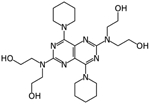
Dipyridamole | Docking in Mpro | IC50 0.53 μM, EC50~0.1 μM | [28] |
