Abstract
DNA double-strand breaks can be repaired through ligation-based pathways (non-homologous end-joining) or replication-based pathways (homologous recombination) in eukaryotic cells. The decisions that govern these outcomes are widely viewed as a competition between factors that recognize DNA ends and physically promote association of factors specific to each pathway, commonly known as “pathway choice”. Here I review recent results in the literature and propose that this decision is better described as a sequential set of binding and end processing events, with non-homologous end joining as the first decision point. Physical association and co-localization of end resection factors with non-homologous end-joining factors suggests that ends are transferred between these complexes, thus the ultimate outcome is not the result of a competition but is more akin to a relay race that is determined by the efficiency of the initial end-joining event and the availability of activated DNA end-processing enzymes.
DNA double-strand breaks (DSBs) are a challenging type of genomic lesion for all cells, as the discontinuity in both strands has the potential for irreversible loss of genetic information and for misrepair events that can generate translocations, insertions, and deletions. The molecular mechanisms of DSB repair have been elucidated over the last several decades starting with genetic studies in bacteria and yeast, followed by in vitro reconstituted assays with purified proteins, single-molecule and structural studies, cell biology and genetics in mammalian systems, and by genome-wide studies facilitated by next-generation sequencing [1–5]. Pathways of DSB repair are generally separated into mechanisms that do not require an intact homologous template (non-homologous end joining (NHEJ), alternative NHEJ, single-strand annealing) and those that utilize a template for replication-driven repair (homologous recombination (HR), break-induced replication)(Figure 1). For in-depth comparisons of these mechanisms see these recent reviews [6–8].
Figure 1. DNA double-strand break repair pathways.
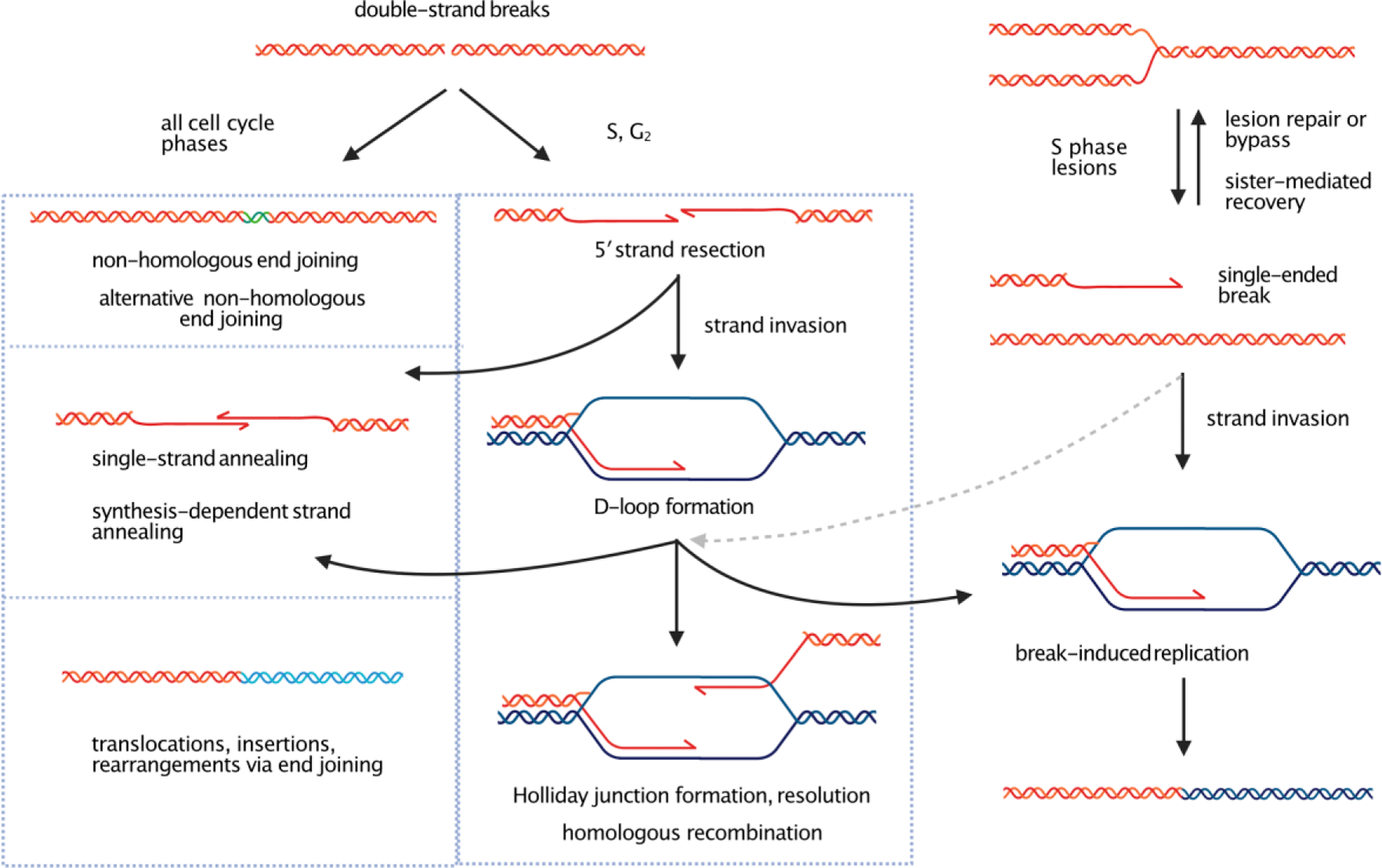
In mammalian cells, repair of DNA double-strand breaks occur via non-homologous end joining during all cell cycle phases and, depending on the structure of the ends, can generate small deletions or insertions (green) at the junction. In S and G2 phases of the cell cycle, resection of 5′ strands is much more efficient than in G1 or G0 phase. Removal of the 5′ strand generates a long, single-stranded 3′ end which is used for strand invasion into unbroken sister chromatids or homologous duplexes. One possible outcome is the formation of a double-Holliday junction and resolution as shown. Resection can also generate intermediates that are used in single-strand annealing, and resection-dependent DNA synthesis can also generate 3′ single-stranded ends that are joined by synthesis-dependent strand annealing. In S phase, lesions in the DNA template can produce single-ended DNA breaks which can be resolved by sister chromatid-mediated strand switching or by strand invasion during break-induced replication from a homologous template as shown (right). Single-ended breaks can also produce misrepair events such as translocations, insertions, and genome rearrangements.
DSB repair outcomes are regulated by resection
One of the key regulation points that affects the outcomes of DSB repair is the resection of 5′ strands at DSBs, a step that is controlled in eukaryotes by cell cycle timing to occur most efficiently and extensively in S and G2 phases [9,10]. The canonical resection process in mammalian cells is initiated by the Mre11-Rad50-Nbs1 (MRN) complex which makes endonucleolytic incisions adjacent to the end, promoted by phosphorylated forms of CtBP-interacting protein (CtIP)[11–14]. The combined actions of MRN and CtIP also promote the binding and activity of the Exo1 and Dna2 nucleases that perform extensive resection of the broken end, degrading hundreds or even thousands of nucleotides depending on the availability of homologous sequences for repair [15–18]. MRN/CtIP-dependent end resection can promote non-templated forms of repair (alternative NHEJ, single-strand annealing) but is considered here primarily as part of the HR pathway.
The outcomes of DSB repair are generally portrayed as a competition between MRN/CtIP and the proteins that recognize breaks and promote NHEJ—the Ku70/Ku80 heterodimer (Ku) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs)(Figure 2). Ku binds to DSB ends and recruits DNA-PKcs, forming the holoenzyme DNA-PK [19] that facilitates the processing of breaks and joining by Ligase IV with the assistance of several accessory factors, including XRCC4, XLF/Cernunnos, Artemis, Cyren/MRI, and polymerases μ and λ [8]. Ku is thought to be the primary end recognition factor for NHEJ and associates very rapidly with laser-induced DNA damage sites in mammalian cells [20,21].
Figure 2. Competition versus Sequential models of DNA double-strand break repair.
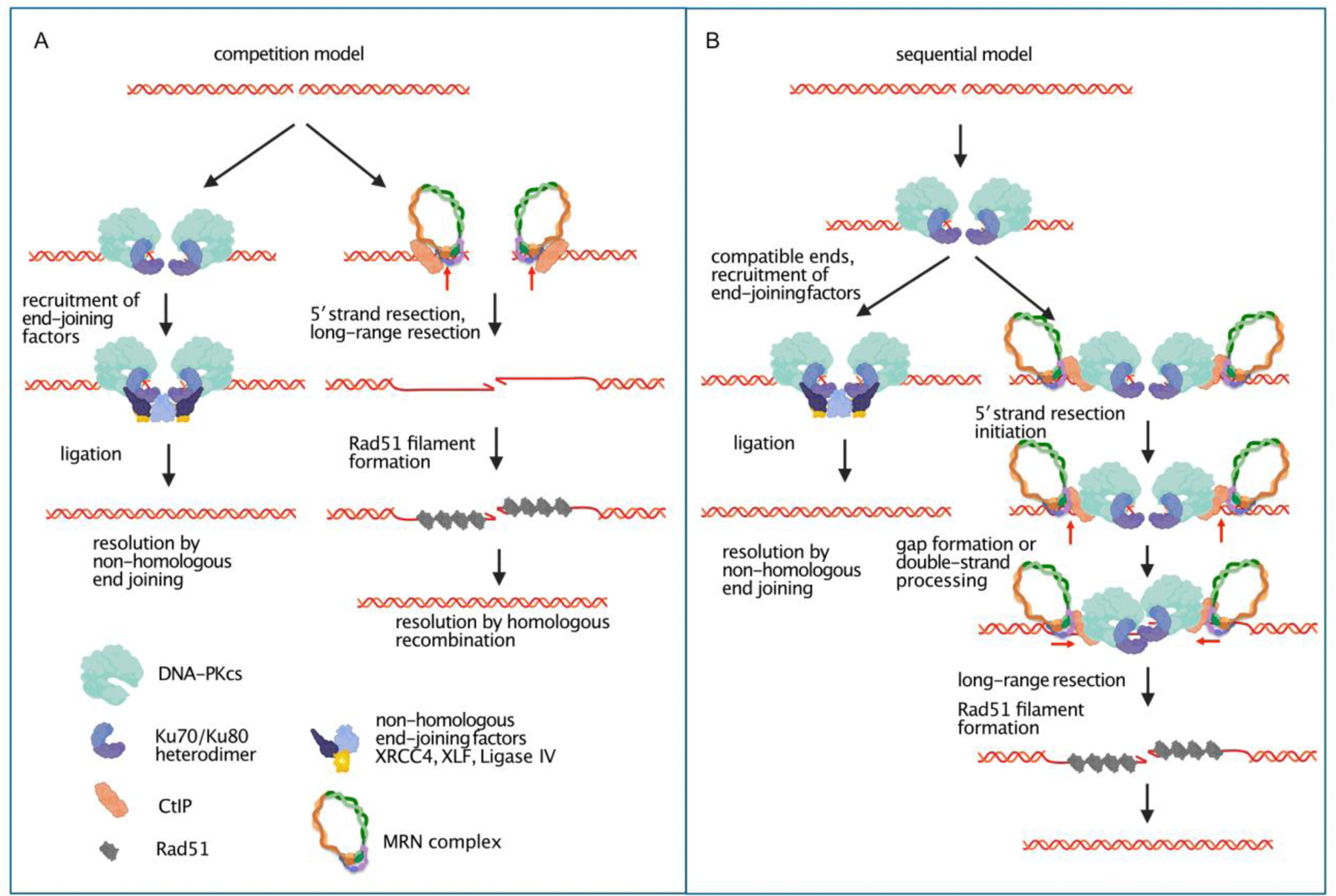
In the simple competition model (A), NHEJ and HR pathways are shown as mutually exclusive, with the initial binding of DNA-PK or MRN/CtIP determining pathway outcomes. Binding of DNA-PK leads to recruitment of NHEJ factors and resolution through ligation, whereas association of MRN and CtIP generate the initial stages of resection followed by long-range resection, RPA association (not shown), Rad51 filament formation, and resolution by HR. For simplicity, DNA-PK is shown only as the holoenzyme (DNA-PKcs with Ku) here. In the sequential model (B), essentially all ends are bound by DNA-PK, with recruitment of NHEJ factors and resolution by ligation occurring when ends are compatible. In this model, failure to resolve (or perhaps failure to productively align) ends by NHEJ leads to the physical association of MRN and CtIP with these DNA-PK-bound breaks, promoting endonucleolytic processing of the ends by MRN, gap formation by 3′ to 5′ exonuclease activity, or double-strand processing. Long-range nuclease recruitment promotes the formation of 3′ single-strands which are subsequently bound by Rad51 and resolved by HR.
The utilization of replication-dependent homologous recombination pathways versus NHEJ differs greatly across organisms, with NHEJ playing a much more prominent role in mammals, coincident with the emergence of DNA-PK [22]. In organisms that predominantly use homologous recombination for DSB repair such as budding and fission yeasts, DNA-PKcs is not present and Ku is expressed at much lower levels compared to mammalian cells [23]. Nevertheless, Ku is still one of the first complexes appearing at a break site in budding yeast [24] and is still an effective block to enzymes other than MRN(X), based on work showing that deletion of Ku subunits allows for DSB 5′ processing in the absence of the MRN complex or CtIP(Ctp1/Sae2)[25–29].
Evidence for competition during pathway choice
A competitive model for DSB repair pathway choice has derived over the years from observations that DNA ends generated in mammalian cells could be repaired by either pathway and that alteration of repair factor levels can skew repair outcomes toward NHEJ or HR [7,30]. Depletion or removal of NHEJ factors reduces the efficiency of NHEJ in rodent and human cells and also increases the relative efficiency of resection or homologous recombination [31–34], indicating that there are compensatory mechanisms that can act to resolve breaks by homology-directed pathways if NHEJ is compromised. NHEJ and HR factors have been shown to associate with DNA ends independently [21,35,36], and in reconstituted biochemical assays in vitro, the presence of Ku inhibits nucleolytic processing of ends [17,37]. These observations are consistent with a competition model in which the initial binding of Ku or MRN dictates whether breaks are resolved by NHEJ or homologous recombination pathways, respectively (Figure 2A).
Despite the attractive simplicity of the competition model, however, other observations are more difficult to reconcile with this view and suggest that we need to reconsider this model.
Observations in support of a sequential model for DSB repair decisions
It is clear in yeast and in mammalian cells that NHEJ factors associate with break sites earlier than HR factors [21,24,35,38,39]. These observations parallel studies of DSB resolution in mammalian cells showing that a fast, NHEJ-dependent joining phase precedes a slower, HR-dependent phase [40,41]. The findings contradict the idea that a subset of breaks is initially designated for resection based on MRN or CtIP association, but are more consistent with a model in which nearly all DSB ends are initially bound by NHEJ-promoting factors. The micromolar levels of DNA-PKcs and Ku in mammals favor this outcome, and human cells contain approximately 50-fold higher levels compared to rodent cells [42], with levels of MRN and CtIP orders of magnitude lower [8].
There are also many observations of co-localization of NHEJ factors with HR factors during the course of DNA repair. Super-resolution microscopy was used to image Ku foci at sites of radiation damage and showed significant co-localization with Mre11 [20], an observation also made in cells exposed to Topoisomerase I inhibitors during replication [43]. In agreement with this finding, purified recombinant MRN complexes and DNA-PK often associate with the same DNA ends in vitro, observed using a single-molecule platform for DNA binding and resection [14,36]. Mre11 was identified as a binding partner for Ku in a recent screen of the Ku interactome [44], reminiscent of Mre11-Ku interactions previously observed in yeast [45]. DNA-PK also phosphorylates the ATM protein kinase, an enzyme that binds to the MRN complex and is recruited via MRN to DNA ends [46], and ATM phosphorylates and regulates DNA-PK [47]. These observations suggest co-occupancy of MRN and Ku as these complexes are the DNA-binding components of ATM and DNA-PKcs, respectively.
Lastly, Ku is enriched at DSB sites in the absence of Mre11 or CtIP(Ctp1/Sae2), including enzymatically induced DSBs in fission yeast and single-ended break sites generated during replication in mammalian cells [25,48]. Importantly, loss of the nuclease activity of Mre11 was shown in both cases to have similar effects on Ku occupancy compared to loss of the protein, therefore Mre11 nuclease activity is inferred to remove the Ku protein from DNA ends in wild-type cells.
In agreement with the idea of MRN/CtIP physically removing Ku from DNA ends, loss of Ku and DNA-PK from ends was observed with the addition of MRN/CtIP using the DNA curtains single-molecule platform [14,36]. In ensemble assays with recombinant human MRN, CtIP, and DNA-PK, the site of MRN cleavage of DNA is approximately 45 nucleotides from the end and absolutely requires the presence of DNA-PKcs as well as Ku [14]. Thus, the presence of DNA-PK promotes the initiating steps of DNA end resection by MRN and CtIP.
In reconstituted assays with purified MRN, CtIP, and DNA-PK, MRN-catalyzed nicking of the DNA on the 5ʹ strand is significantly more efficient than simultaneous cuts on both strands, suggesting that single-strand processing is possible and perhaps even a preferred mechanism. Results in fission yeast showed that limited resection of Ku-bound DSBs could occur under conditions of Mre11 nuclease deficiency or Ctp1 loss, such that RPA-bound single-stranded DNA is produced adjacent to a (presumably) double-stranded DNA end bound by Ku [25]. This type of single-strand gap formation was initially shown to occur via the endonuclease and exonuclease activities of the MRX complex in budding yeast at sites of Spo11 covalent DSBs [49], consistent with the idea that this is an evolutionarily conserved function of the complex.
Taken together, these observations lead to a view of DSB repair decision-making in mammalian cells that is fundamentally different from a simple competition model and has previously been proposed in varying ways as a NHEJ-first scenario [7–9,50]. In this sequential model (Figure 2B), NHEJ-related factors associate with DNA ends first and promote end-joining if the ends are compatible. If NHEJ is unsuccessful, DNA-PK complexes become long-lived on the ends, promoting the binding and processing step of resection by MRN and CtIP. Since DNA-PK is an essential component of this reaction, the process is necessarily a physical transfer from NHEJ to HR pathways that is enforced by the fact that MRN in collaboration with phosphorylated CtIP appears to be the only “key” that opens the “lock” generated by DNA-PK on ends. Recent mechanistic modeling of DSB repair using a meta-analysis of experimental data also supports the idea of a “entwined relationship” between NHEJ and HR rather than simple competition scenarios [51]. Observations that correlate the complexity of DSB ends with the efficiency of resection [52] also are consistent with this model, since the presence of adducts or unligatable ends reduces the likelihood of successful NHEJ.
A sequential model may also be attractive as a framework for understanding DNA end processing events during NHEJ as well as alternative NHEJ (alt-NHEJ), where limited deletions of DNA at the ends (<20 nt) facilitate end joining, often utilizing microhomologies at the breakpoints [8]. The Artemis endonuclease is generally considered to be the primary nuclease facilitating NHEJ, although the MRN complex and ATM have also been shown to be required for processing and joining of DNA ends through an Artemis-dependent subpathway of NHEJ that is responsible for repair of approximately 10% of radiation-induced DSBs [53]. The MRN complex was also found to bind to and promote the activity of ligase III/XRCC1, a complex that functions in alt-NHEJ when the major NHEJ pathway is inactive [54]. It is conceivable that these pathways may be promoted by MRN at DNA-PK-bound ends when classical NHEJ is unsuccessful.
The role of DNA-PKcs catalytic activity
Unlike ATM, DNA-PK has relatively few verified protein targets in mammalian cells; however, DNA-PKcs autophosphorylation on conserved clusters of sites and phosphorylation of Ku70 definitely occur in cells and have important consequences [19,55]. These phosphorylation events lead to conformational changes that are essential for the process of NHEJ but also ultimately result in the disassociation of DNA-PKcs and Ku from DNA [56–61]. Inhibition of DNA-PK kinase activity with chemical inhibitors promotes higher levels of DNA end resection and homologous recombination [33,62,63], consistent with the idea that blocking release of DNA-PK from DNA leads to a stable, perhaps irreversible, complex that promotes MRN/CtIP activity. Other results using catalytic mutants of DNA-PKcs also show that the presence of the mutant enzyme induces levels of HR that are significantly higher than in the absence of DNA-PKcs [64]. This is an important finding since NHEJ is equivalently blocked in both the absence of DNA-PKcs and with expression of the mutant kinase yet higher HR is only seen with the protein present, indicating an important pro-resection role for the catalytically inactive protein. It should be noted, however, that other studies utilizing different mutants and other chemical inhibitors showed contradictory results [52,65,66], so the picture is not entirely clear.
Catalytic mutants of the DNA damage-related PI-3-like kinases ATM, DNA-PKcs, and ATR generally have very different biological effects compared to complete loss of the enzymes [67]. DNA-PKcs catalytic mutants for instance cannot sustain embryonic viability in mice, despite the mild, immune system-specific phenotype of a DNA-PKcs deletion in the same organism, and deletion of Ku from DNA-PKcs kinase-deficient mice rescues this lethality [59].
Conclusions and further questions
The recent insights into mechanisms of DSB pathway decisions in mammalian cells suggest a sequential hand-off model as an alternative to the simple competition model usually depicted in the literature (Figure 2). However, many questions remain about the details of this process as it occurs in the context of living cells.
What are the patterns of single-strand or double-strand processing by MRN at sites of DNA-PK binding and how do these change with cell cycle phase? A modified ChIP assay was developed to isolate small fragments of DNA bound by DNA-PK that are released from chromatin by MRN (“Gentle Lysis and Size Selection”“ or GLASS-ChIP)[14], but it is not yet clear what the patterns are genome-wide and how prevalent or extensive gap formation is.
What happens in organisms lacking DNA-PKcs? Phylogenetic analysis of DNA-PKcs evolution shows that there are orthologs in a broad range of eukaryotes [68], although several commonly used model organisms, e.g. S. cerevisiae, S. pombe, D. melanogaster, C. elegans, lack an obvious DNA-PKcs enzyme. Ku antagonizes resection and HR pathways in budding and fission yeasts as described above, and in vitro experiments with budding yeast MRX have shown that Ku as well as other proteins can act as protein blocks that promote Mre11 endonucleolytic activity [69]. Whether this block occurs in the same way in other organisms is not clear, and whether other proteins functionally substitute for DNA-PKcs in these cases is not known. The relationship between the MRX complex and NHEJ factors is different in budding yeast compared to mammalian cells since MRX is required for efficient NHEJ in S. cerevisiae [45,70–73]. This physical association and functional relationship between MRX and NHEJ factors in budding yeast even led to an early suggestion that MRX may play the role of DNA-PKcs in yeast [72]. It is possible that Mre11-Rad50 complexes have evolved to work cooperatively and in direct association with Ku, consistent with the observation that Rad50 (SbcC) acts in the same repair pathway as bacterial Ku in B. subtilis [74]. From this point of view, it may be useful to entertain the idea of Ku and MRN(X) as a functional unit, with repair outcomes determined by levels of each complex and the structure of the ends.
An important question that remains to be answered is: what determines the timing of MRN/CtIP-mediated processing of DNA-PK-bound ends? The appearance of “toxic NHEJ” events at single-ended double-strand breaks and observations of enhanced NHEJ in ATM-deficient cells suggests that ATM plays a major role in orchestrating end processing and restricting NHEJ [75,76]. The critical ATM-dependent events are not yet clear, however, and it is not known if there is a conformational change in DNA-PK or other proteins on the ends that initiate the MRN processing. The phosphorylation of DNA-PKcs by ATM [47] could conceivably be part of this regulation.
Lastly, what is the relationship between transcription and the interplay between MRN and Ku at DNA ends? Many recent studies suggest that the formation of RNA-DNA hybrids have important effects on resection and HR efficiency and that transcription affects the occupancy of MRN at DNA ends [77–84]. MRN was also recently shown to associate directly with sites of RNA polymerase II binding in human cells [85].We know that transcription generally promotes resection and HR but the mechanisms and involvement of MRN in this process are not fully elucidated.
Answering these questions, attaining structures of the relevant multi-component complexes on DNA, and determining how the many chromatin-bound factors that influence MRN and Ku function are acting in these pathways will be essential for a mechanistic understanding of DSB repair decisions.
Acknowledgements
Thanks to Jeremy Stark for helpful comments on the manuscript. This work was supported in part by the National Institutes of Health (P01CA092584) and the Cancer Research and Prevention Institute of Texas (RP200254).
Funding
No funding was received for this work
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
No conflict of interest exists
References
- 1.Falk M, Hausmann M: A Paradigm Revolution or Just Better Resolution—Will Newly Emerging Superresolution Techniques Identify Chromatin Architecture as a Key Factor in Radiation-Induced DNA Damage and Repair Regulation? Cancers 2020, 13:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J: The DNA-damage response in human biology and disease. Nature 2009, 461:1071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowalczykowski SC: An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb Perspect Biol 2015, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symington LS, Gautier J: Double-strand break end resection and repair pathway choice. Annual review of genetics 2011, 45:247–71. [DOI] [PubMed] [Google Scholar]
- 5.Vítor AC, Huertas P, Legube G, de Almeida SF: Studying DNA Double-Strand Break Repair: An Ever-Growing Toolbox. Front Mol Biosci 2020, 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceccaldi R, Rondinelli B, D’Andrea AD: Repair Pathway Choices and Consequences at the Double-Strand Break. Trends in Cell Biology 2016, 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scully R, Panday A, Elango R, Willis NA: DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol 2019, 20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang HHY, Pannunzio NR, Adachi N, Lieber MR: Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nature Reviews Molecular Cell Biology 2017, 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsuki Y, Jeggo PA, Uchihara Y, Takata M, Shibata A: DNA double-strand break end resection: a critical relay point for determining the pathway of repair and signaling. GENOME INSTAB DIS 2020, 1:155–171. [Google Scholar]
- 10.Symington LS: Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 2016, 51:195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anand R, Ranjha L, Cannavo E, Cejka P: Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell 2016, 64:940–950. [DOI] [PubMed] [Google Scholar]
- 12.Cannavo E, Cejka P: Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 2014, 514:122–5. [DOI] [PubMed] [Google Scholar]
- 13.Deshpande RA, Lee JH, Arora S, Paull TT: Nbs1 Converts the Human Mre11/Rad50 Nuclease Complex into an Endo/Exonuclease Machine Specific for Protein-DNA Adducts. Mol Cell 2016, 64:593–606. Deshpande et al show evidence for DNA-PK-promoted MRN/CtIP processing of DNA ends in vitro and in human cells. Reconstitution of MRN endonucleolytic cleavage of DNA with purified components shows that cutting is dependent on DNA-PKcs, Ku, and phosphorylated CtIP. A modified ChIP assay (GLASS-ChIP) shows that MRN-induced, DNA-PK-bound fragments are produced at double-strand break sites in human cells. [DOI] [PubMed] [Google Scholar]
- 14.Deshpande RA, Myler LR, Soniat MM, Makharashvili N, Lee L, Lees-Miller SP, Finkelstein IJ, Paull TT: DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci Adv 2020, 6:eaay0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC: BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 2011, 25:350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. : Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 2010, 467:108–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE: Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 2010, 29:3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syed A, Tainer JA: The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu Rev Biochem 2018, 87:263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jette N, Lees-Miller SP: The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Progress in biophysics and molecular biology 2015, 117:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britton S, Coates J, Jackson SP: A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. The Journal of cell biology 2013, 202:579–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kochan JA, Desclos ECB, Bosch R, Meister L, Vriend LEM, van Attikum H, Krawczyk PM: Meta-analysis of DNA double-strand break response kinetics. Nucleic Acids Research 2017, 45:12625–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrivastav M, De Haro LP, Nickoloff JA: Regulation of DNA double-strand break repair pathway choice. Cell research 2008, 18:134–47. [DOI] [PubMed] [Google Scholar]
- 23.Fell VL, Schild-Poulter C: The Ku heterodimer: function in DNA repair and beyond. Mutat Res Rev Mutat Res 2015, 763:15–29. [DOI] [PubMed] [Google Scholar]
- 24.Wu D, Topper LM, Wilson TE: Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 2008, 178:1237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langerak P, Mejia-Ramirez E, Limbo O, Russell P: Release of Ku and MRN from DNA Ends by Mre11 Nuclease Activity and Ctp1 Is Required for Homologous Recombination Repair of Double-Strand Breaks. PLoS genetics 2011, 7:e1002271. This study in fission yeast showed that Mre11 nuclease activity and Ctp1 (S. pombe ortholog of CtIP) are required for dissociation of Ku from DNA ends. This dissociation is proposed to be a critical step for RPA loading and subsequent homologous recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P: Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 2007, 28:134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mimitou EP, Symington LS: Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 2010, 29:3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, et al. : Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 2003, 23:5186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasko BM, Holland CL, Resnick MA, Lewis LK: Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair 2009, 8:162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han J, Huang J: DNA double-strand break repair pathway choice: the fork in the road. GENOME INSTAB DIS 2020, 1:10–19. [Google Scholar]
- 31.Pierce AJ, Hu P, Han M, Ellis N, Jasin M: Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 2001, 15:3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M: Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 2004, 24:9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Paull TT: DNA-dependent Protein Kinase Regulates DNA End Resection in Concert with Mre11-Rad50-Nbs1 (MRN) and Ataxia Telangiectasia-mutated (ATM). J Biol Chem 2013, 288:37112–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Caron P, Legube G, Paull TT: Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic acids research 2014, 42:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J-S, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AMR, Yokomori K: Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. The Journal of Cell Biology 2005, 170:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myler LR, Gallardo IF, Soniat MM, Deshpande RA, Gonzalez XB, Kim Y, Paull TT, Finkelstein IJ: Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol Cell 2017, 67:891–898 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J, Lee K-J, Davis AJ, Chen DJ: Human Ku70/80 Protein Blocks Exonuclease 1-mediated DNA Resection in the Presence of Human Mre11 or Mre11/Rad50 Protein Complex*. Journal of Biological Chemistry 2012, 287:4936–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones CE, Forsburg SL: Monitoring Schizosaccharomyces pombe genome stress by visualizing end-binding protein Ku. Biology Open 2021, 10:bio054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang G, Liu C, Chen S-H, Kassab MA, Hoff JD, Walter NG, Yu X: Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res 2018, 46:3446–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA: gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell cycle 2010, 9:662–9. [DOI] [PubMed] [Google Scholar]
- 41.Mao Z, Bozzella M, Seluanov A, Gorbunova V: Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008, 7:1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finnie NJ, Gottlieb TM, Blunt T, Jeggo PA, Jackson SP: DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proceedings of the National Academy of Sciences 1995, 92:320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whelan DR, Lee WTC, Marks F, Kong YT, Yin Y, Rothenberg E: Super-resolution visualization of distinct stalled and broken replication fork structures. PLoS Genet 2020, 16:e1009256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abbasi S, Schild-Poulter C: Mapping the Ku Interactome Using Proximity-Dependent Biotin Identification in Human Cells. J Proteome Res 2019, 18:1064–1077. [DOI] [PubMed] [Google Scholar]
- 45.Palmbos PL, Daley JM, Wilson TE: Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol Cell Biol 2005, 25:10782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y, Lee JH, Jiang W, Crowe JL, Zha S, Paull TT: Regulation of the DNA Damage Response by DNA-PKcs Inhibitory Phosphorylation of ATM. Mol Cell 2017, 65:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ: Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem 2007, 282:6582–7. [DOI] [PubMed] [Google Scholar]
- 48.Chanut P, Britton S, Coates J, Jackson SP, Calsou P: Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nature communications 2016, 7:12889. This study investigated single-ended double-strand breaks created during S phase by Top1 poisons and found that ATM-mediated phosphorylation of CtIP as well as Mre11 endonuclease activity are required to block Ku accumulation at these ends. Mre11 exo- and endonuclease activity as well as CtIP nuclease activity are proposed to act in a coordinated way to remove Ku in this scenario. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia V, Phelps SE, Gray S, Neale MJ: Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 2011, 479:241–4. This study in budding yeast was the first to show that Spo11-linked DNA ends are processed by Mre11 such that endonuclease activity followed by exonucleolytic degradation generates a gapped intermediate that ultimately facilitates homologous recombination during meiosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shibata A: Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2017, 803–805:51–55. [DOI] [PubMed] [Google Scholar]
- 51.Ingram SP, Warmenhoven JW, Henthorn NT, Smith EAK, Chadwick AL, Burnet NG, Mackay RI, Kirkby NF, Kirkby KJ, Merchant MJ: Mechanistic modelling supports entwined rather than exclusively competitive DNA double-strand break repair pathway. Sci Rep 2019, 9:6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Löbrich M, et al. : Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J 2011, 30:1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. : A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 2004, 16:715–24. [DOI] [PubMed] [Google Scholar]
- 54.Della-Maria J, Zhou Y, Tsai M-S, Kuhnlein J, Carney JP, Paull TT, Tomkinson AE: Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem 2011, 286:33845–33853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee KJ, Saha J, Sun J, Fattah KR, Wang SC, Jakob B, Chi L, Wang SY, Taucher-Scholz G, Davis AJ, et al. : Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic acids research 2016, 44:1732–45. Lee and colleagues identify phosphorylation sites for DNA-PKcs in the Ku70 protein and show that these modifications are important for release of Ku from DNA ends and for the initiation of homologous recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP: Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic acids research 2004, 32:4351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ: Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 2002, 16:2333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K: Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 2003, 23:5836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, Lee BJ, Dubois RL, Liu C, Yu X, et al. : Differential Phosphorylation of DNA-PKcs Regulates the Interplay between End-Processing and End-Ligation during Nonhomologous End-Joining. Molecular Cell 2015, 58:172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddy YV, Ding Q, Lees-Miller SP, Meek K, Ramsden DA: Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J Biol Chem 2004, 279:39408–13. [DOI] [PubMed] [Google Scholar]
- 61.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ: Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. The Journal of cell biology 2007, 177:219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robert F, Barbeau M, Éthier S, Dostie J, Pelletier J: Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 2015, 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C, Tang H, Geng A, Dai B, Zhang H, Sun X, Chen Y, Qiao Z, Zhu H, Yang J, et al. : Rational combination therapy for hepatocellular carcinoma with PARP1 and DNA-PK inhibitors. Proc Natl Acad Sci USA 2020, 117:26356–26365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shrivastav M, Miller CA, De Haro LP, Durant ST, Chen BPC, Chen DJ, Nickoloff JA: DNA-PKcs and ATM co-regulate DNA double-strand break repair. DNA Repair 2009, 8:920–929. This study investigates the role of DNA-PKcs catalytic activity in pathway choice. They show that kinase-deficient forms of DNA-PKcs promote homologous recombination, in contrast to cells lacking DNA-PKcs, suggesting that the inactive kinase likely plays a regulatory role in this decision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen C, Halbrook J, Nickoloff JA: Interactive competition between homologous recombination and non-homologous end joining. Molecular cancer research : MCR 2003, 1:913–20. [PubMed] [Google Scholar]
- 66.Convery E, Shin EK, Ding Q, Wang W, Douglas P, Davis LS, Nickoloff JA, Lees-Miller SP, Meek K: Inhibition of homologous recombination by variants of the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs). Proceedings of the National Academy of Sciences 2005, 102:1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menolfi D, Zha S: ATM, ATR and DNA-PKcs kinases—the lessons from the mouse models: inhibition ≠ deletion. Cell Biosci 2020, 10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lees-Miller JP, Cobban A, Katsonis P, Bacolla A, Tsutakawa SE, Hammel M, Meek K, Anderson DW, Lichtarge O, Tainer JA, et al. : Uncovering DNA-PKcs ancient phylogeny, unique sequence motifs and insights for human disease. Progress in Biophysics and Molecular Biology 2020, doi: 10.1016/j.pbiomolbio.2020.09.010. This study investigates the evolution of DNA-PKcs and finds that the protein is surprisingly widespread in invertebrates, fungi, plants, and protists, in addition to mammalian organisms. Conserved motifs are identified that are specific to DNA-PKcs and are likely to be functionally important for end joining in eukaryotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang W, Daley JM, Kwon Y, Krasner DS, Sung P: Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev 2017, 31:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE: Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol 2007, 14:639–46. [DOI] [PubMed] [Google Scholar]
- 71.Milne GT, Jin S, Shannon KB, Weaver DT: Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol 1996, 16:4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE: Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 2001, 8:1105–15. [DOI] [PubMed] [Google Scholar]
- 73.Matsuzaki K, Shinohara A, Shinohara M: Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics 2008, 179:213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mascarenhas J, Sanchez H, Tadesse S, Kidane D, Krisnamurthy M, Alonso JC, Graumann PL: Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair. BMC Mol Biol 2006, 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balmus G, Pilger D, Coates J, Demir M, Sczaniecka-Clift M, Barros AC, Woods M, Fu B, Yang F, Chen E, et al. : ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat Commun 2019, 10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhargava R, Carson CR, Lee G, Stark JM: Contribution of canonical nonhomologous end joining to chromosomal rearrangements is enhanced by ATM kinase deficiency. Proc Natl Acad Sci U S A 2017, 114:728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crossley MP, Bocek M, Cimprich KA: R-Loops as Cellular Regulators and Genomic Threats. Molecular Cell 2019, 73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Domingo‐ Prim J, Bonath F, Visa N: RNA at DNA Double‐ Strand Breaks: The Challenge of Dealing with DNA:RNA Hybrids. BioEssays 2020, 42:1900225. [DOI] [PubMed] [Google Scholar]
- 79.Paull TT: RNA-DNA hybrids and the convergence with DNA repair. Crit Rev Biochem Mol Biol 2019, 54:371–384. [DOI] [PubMed] [Google Scholar]
- 80.Sharma S, Anand R, Zhang X, Francia S, Michelini F, Galbiati A, Williams H, Ronato DA, Masson J-Y, Rothenberg E, et al. : MRE11-RAD50-NBS1 Complex Is Sufficient to Promote Transcription by RNA Polymerase II at Double-Strand Breaks by Melting DNA Ends. Cell Reports 2021, 34:108565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu S, Hua Y, Wang J, Li L, Yuan J, Zhang B, Wang Z, Ji J, Kong D: RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell 2021, 184:1314–1329.e10. [DOI] [PubMed] [Google Scholar]
- 82.Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, et al. : Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 2014, 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yasuhara T, Kato R, Hagiwara Y, Shiotani B, Yamauchi M, Nakada S, Shibata A, Miyagawa K: Human Rad52 Promotes XPG-Mediated R-loop Processing to Initiate Transcription-Associated Homologous Recombination Repair. Cell 2018, 175:558–570.e11. [DOI] [PubMed] [Google Scholar]
- 84.Ouyang J, Yadav T, Zhang J-M, Yang H, Rheinbay E, Guo H, Haber DA, Lan L, Zou L: RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 2021, 594:283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salifou K, Burnard C, Basavarajaiah P, Grasso G, Helsmoortel M, Mac V, Depierre D, Franckhauser C, Beyne E, Contreras X, et al. : Chromatin-associated MRN complex protects highly transcribing genes from genomic instability. Sci Adv 2021, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]


