Figure 1. EditPredict model schema. The two left panels with light grey shading correspond to the training (top) and application (bottom) processes, respectively.
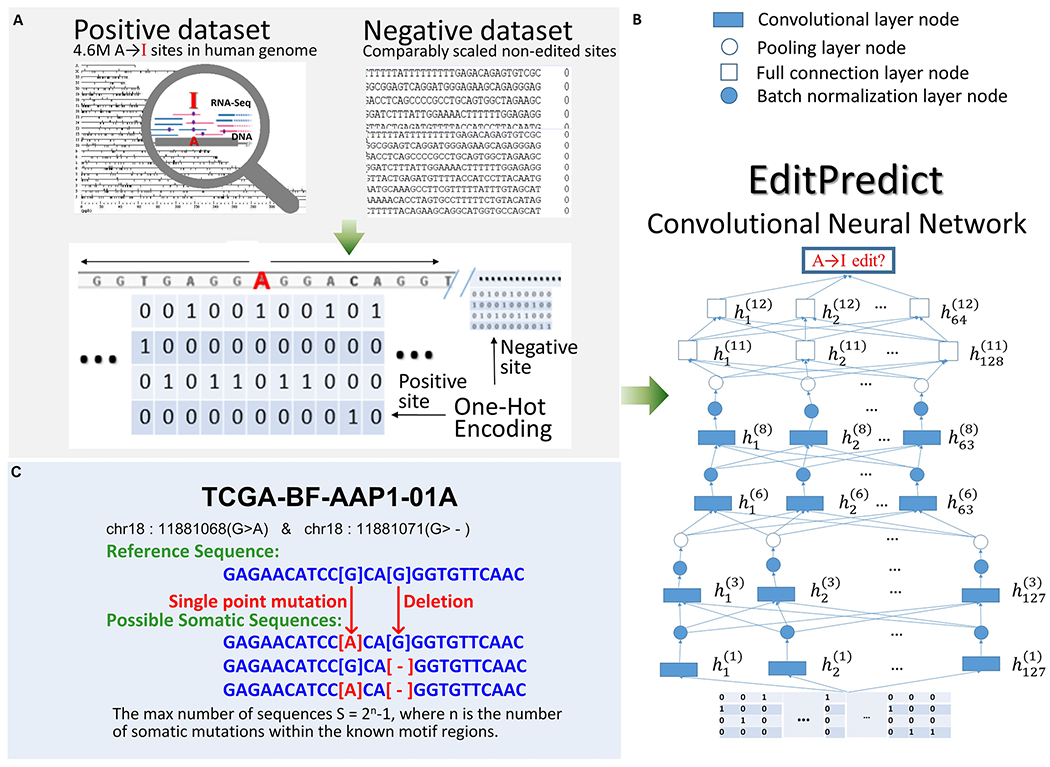
A. Approximately, flanking sequences of ~4.6 million known RNA editing sites were extracted as the positive set, and matched negative sites were constructed. One-hot encoding for a segment of 11 nucleotides was illustrated. B. The CNN architecture handles bi-directional long flanking sequences of potential A-to-I editing sites. The number of layers and the number of neurons per layer are depicted. The convolution process has been omitted for clarity. C. An example of a personalized genome approach. Multiple flanking sequences for a real subject may be generated based on mutation and INDELs.
