Figure 2. Performance of various models of RNA editing prediction out of ten-fold cross-validation.
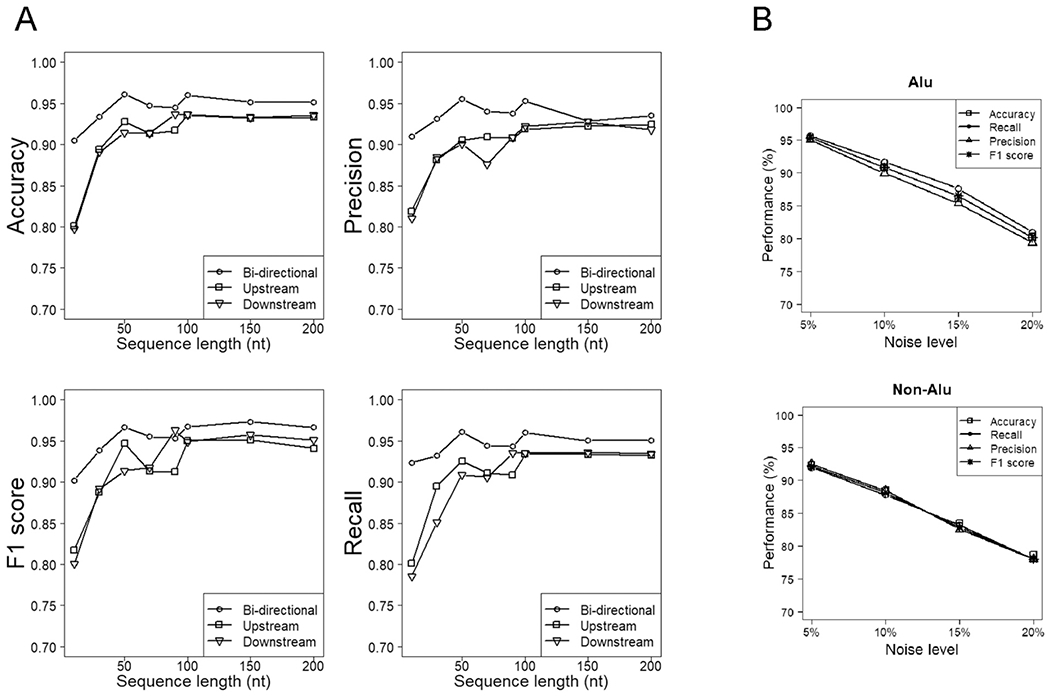
A, Performance of the overall models (with Alu and non-Alu RNA editing sites combined) of various directional modes and handling sequence lengths. “Upstream” and “downstream” refer to sequences adjoining the concerned sites in only one direction, while “both sides” refer to sequences extended from the central concerned sites bidirectionally. Compared to the uni-directional models, the corresponding bi-directional models deal with sequences of a roughly doubled length, although they are depicted with the same series of length parameters for illustrative symmetry. B, Performance of the Alu/non-Alu models trained on perturbed training datasets, where a percentage of positive datapoints were replaced with random negative datapoints.
