Figure 3. A-to-I RNA editing associations with Alu elements.
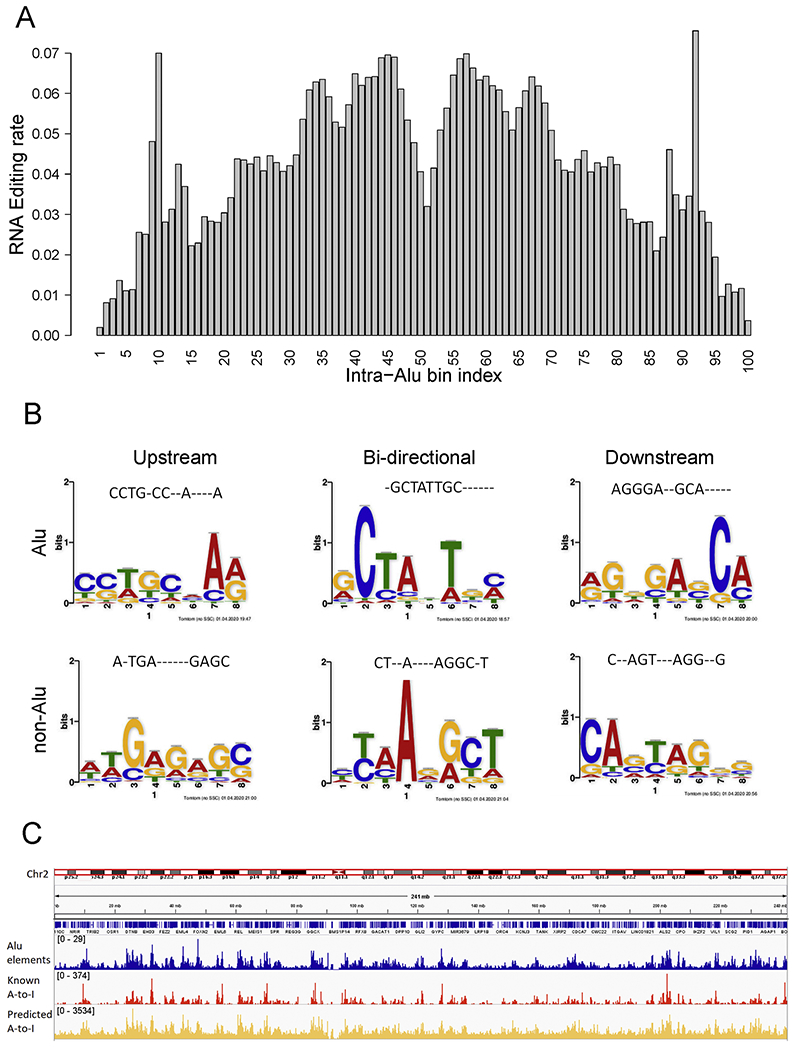
A) A-to-I RNA editing site distribution over the span of an Alu segment (normalized to 100 bins). Hotspot of A-to-I RNA editing can be observed near the boundary (10th & 92th bins), and the other peak is near the center point (45th & 57th bins). The precise boundary sites have the least likelihood of editing, and the absolute center has a moderate level of editing. This figure used data only from the positive strand perspective, thus producing a symmetry effect. B) Motifs from upstream, downstream and bi-directional models filters with highest weight. Each sequence logo is converted to an oligo nucleotide sequence by taking the nucleotide of the highest probability, and the alignment of the motif sequence with the best-hitting previous RNA editing motif is displayed on the top. C) Overlay of Alu elements, known A-to-I RNA editing sites, and predicted A-to-I RNA editing sites across chromosome 2. Similar peaks location can be observed.
