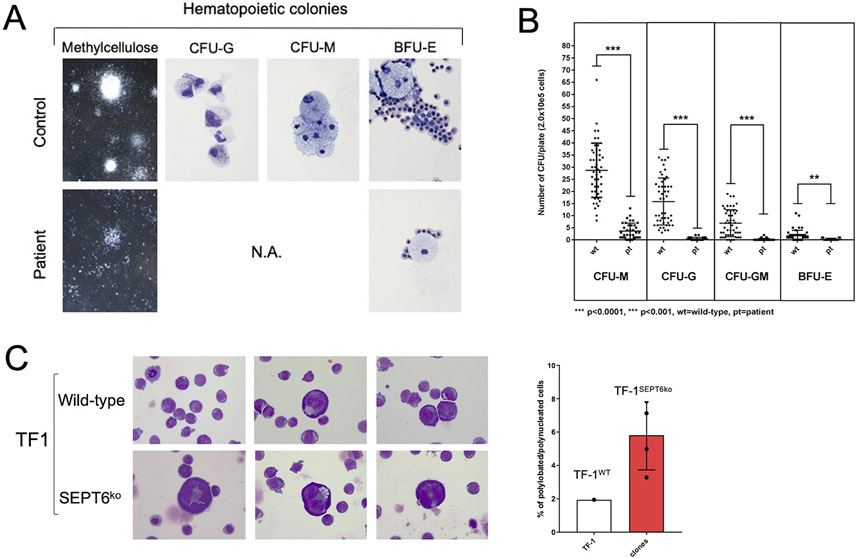
Figure 4: Derivation of hematopoietic progenitors from patient-iPSC colonies phenocopies the hematological findings of SEPT6-associated congenital MDS, and the genetic knock-out of SEPT6 in hematopoietic cell lines reveals a propensity to multinuclearity.
Embryoid bodies (EB) were generated from patient and healthy control iPSCs (Supplementary Figure 2A), which were morphologically comparable. From EBs, hematopoietic progenitor cells (HPCs) were extracted, plated into differentiation-cytokine containing methycellulose (2.0x10e5 cells per 5cm diameter dish), and analyzed at maximal expansion of colony-forming units (CFUs). CFUs were scored by standard microscopical means following morphological guidelines into granulocyte (G), monocyte (M), granulocyte-monocyte (GM) and erythroid (E) categories and counted twice. Larger colonies were aspirated by pipetting and cytospins stained by MGG. Patient-derived CFUs were markedly reduced in numbers and size at maximal expansion in methylcellulose when compared to CFUs from control iPSCs (panel A, left). We generated myeloid lineage progenitors (CFU-G, -GM, -M, -E) from patient-derived iPSC clones at good efficiency, and cellular morphology confirmed to be correct after MGG staining of cytospins of larger representative colonies (panel A, right, 100x magnification). This was not the case for HPCs generated from patient-derived iPSC clones, where colonies were too small to allow morphological confirmation. To numerically quantify the defect, we counted CFUs and observed a defective granulocyte/monocyte development of CFUs from patient-derived iPSCs. We measured a differential effect on the myeloid/granulocyte lineage compared to the erythroid lineage with a reduction by 36-fold in CFU-G, 46-fold CFU-GM, 8-fold in CFU-M, and 6-fold in BFU-E colony output in the patient derived HPC when compared to controls (panel B). Data is from n=3 non-synchronous experiments, pooling iPSC clones (c5 and c8 for patient, c1 and c6 for control). Statistical significance was calculated by Student t-testing. Wt= wild type controls, Pt= patient derived cells, ****p<0.0001, ***p<0.001.
After attempting to knock-in/out SEPT6 in iPSCs and human granulocytic/myeloid cell lines, which showed an intolerance to SEPT6 insufficiency, we genetically edited the erythroleukemic cell line TF-1 by CRISPR/Cas9 and achieved a complete SEPT6 knock-out in pooled cells which remained stable in multiple generations of single cell clones (Supplementary Figure 5A-B). After culture displaying no difference in proliferative dynamics, we noted giant cells with multinucleation in SEPT6 knock-out clones, when cytospins were stained by MGG, while this could not be seen in wild-type TF-1 controls (panel C left, 40x magnification). To address this observation numerically, we counted nucleated cells by categories (mono-, bi- polynucleation), and identified an increased proportion of larger, multinucleated/lobated cells in SEPT6 knock-out clones. Data is from n>1000 for each SEPT6 ko clone and wild-type, one experiment representative of three. When SEPT6 knock-out clone counts were pooled and frequency of multinucleated/lobated cells was compared with wild-type TF-1 cells, a 3-fold increase was observed (panel C right). This population of cells was insufficient to cause differences in DNA content and cell cycle as assayed by flow cytometry after PI/7-AAD staining.