Figure 4. Jpx selectively controls a subset of developmentally sensitive CTCF motifs.
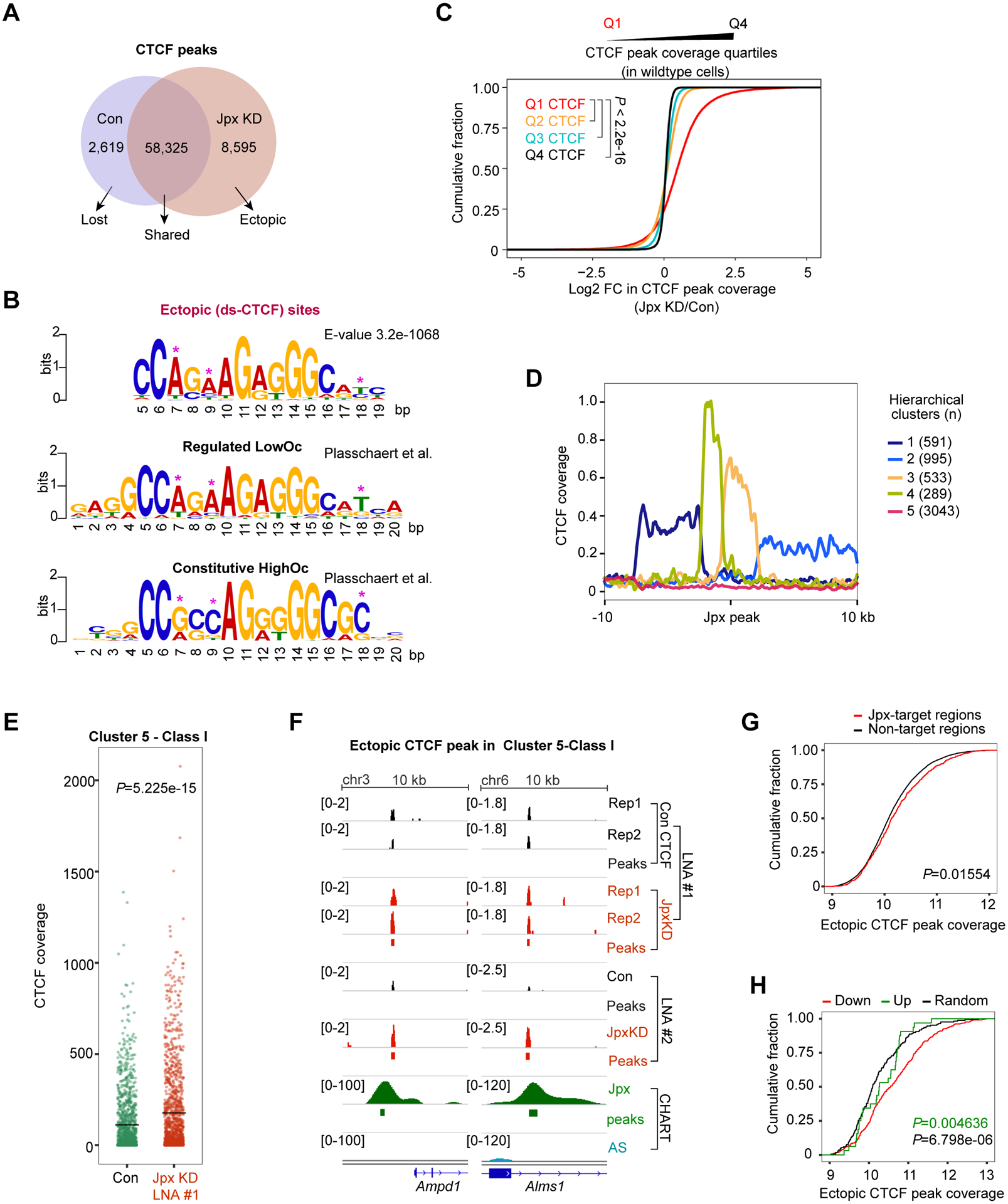
(A) Venn diagram representing the number of overlapping or exclusive CTCF sites in control and Jpx-depleted d7 ES cells. CTCF peaks common to two biological replicates were used.
(B) Logograms of sequence motifs for 8,595 ectopic CTCF peaks described in (A). LowOc and HighOc sites as defined by Plasschaert et al.
(C) CDP comparing log2 FC in CTCF peak coverage over Q1, Q2, Q3 and Q4 CTCF quartiles. CTCF sites were divided into quartiles based on peak coverage. P-values, the KS test.
(D) Five hierarchical clusters defined by CTCF’s relationship to Jpx peaks in d7 ES cells. See also Fig. S6A.
(E) CTCF coverage values calculated over Cluster 5-Class I genes in control vs. Jpx LNA#1 cells. See also Fig. S6E. Black crossbar, mean. P-value, Wilcoxon ranked sum test.
(F) Cluster 5-Class I: Increased CTCF coverages following Jpx depletion.
(G) CDP comparing log2 coverage of ectopic CTCF peaks between Jpx-targets and non-targets (20 kb bins). P-value, KS test.
(H) CDP showing log2 coverage of ectopic CTCF peaks for down-DEGs (n=738), up-DEGs (n=162) and randomly selected non-DEGs (n=900). P-values, KS test: Green, down vs. up. Black, down vs. random.
