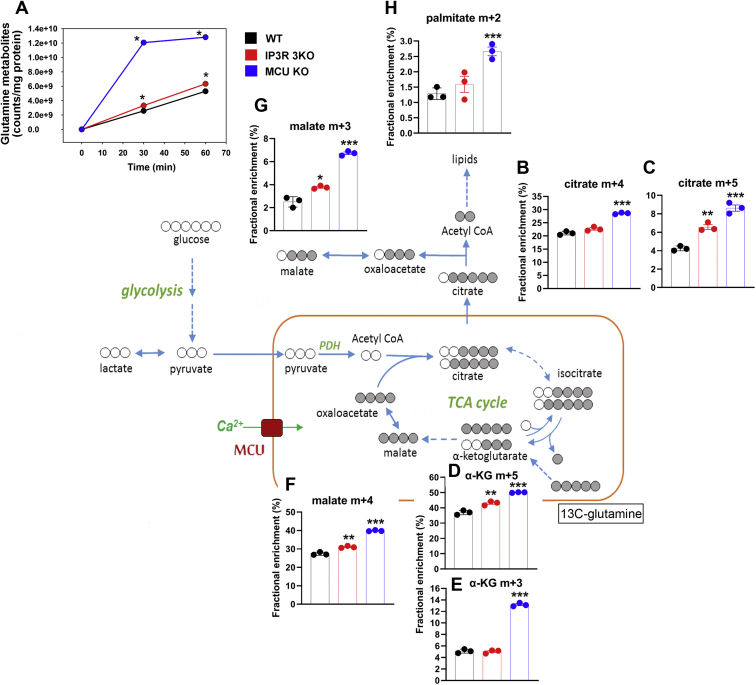
Figure 6.
[U–13C]-glutamine tracer analysis WT, IP3R TKO, and MCU KO cells. Cells were washed in glutamine-free DMEM and then incubated in a DMEM containing 5 mM uniformly labeled 13C-glutamine for 30 or 60 min. Samples were prepared for LC–MS/MS as described in the “Experimental procedures” section. A, the sum of counts appearing as glutamate, α-ketoglutarate, aspartate, citrate/isocitrate, and cis-aconitate was taken as a measure of glutamine metabolism. The data were normalized to protein. The error bars were smaller than symbol size. Only the 60 min data for selected metabolites are shown in (B–H). Data are the fractional enrichment of the indicated isotopolog given as the mean ± SEM of three separate plates for each condition. A simplified cartoon of the flow of labeled carbon (shaded gray) is shown with steps involving multiple enzymes being indicated by dotted arrows. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001, one-way ANOVA followed by multiple comparison analysis using Dunnett’s test. DMEM, Dulbecco's modified Eagle's medium; IP3R, inositol trisphosphate receptor; MCU, mitochondrial Ca2+ uniporter; TKO, triple KO.