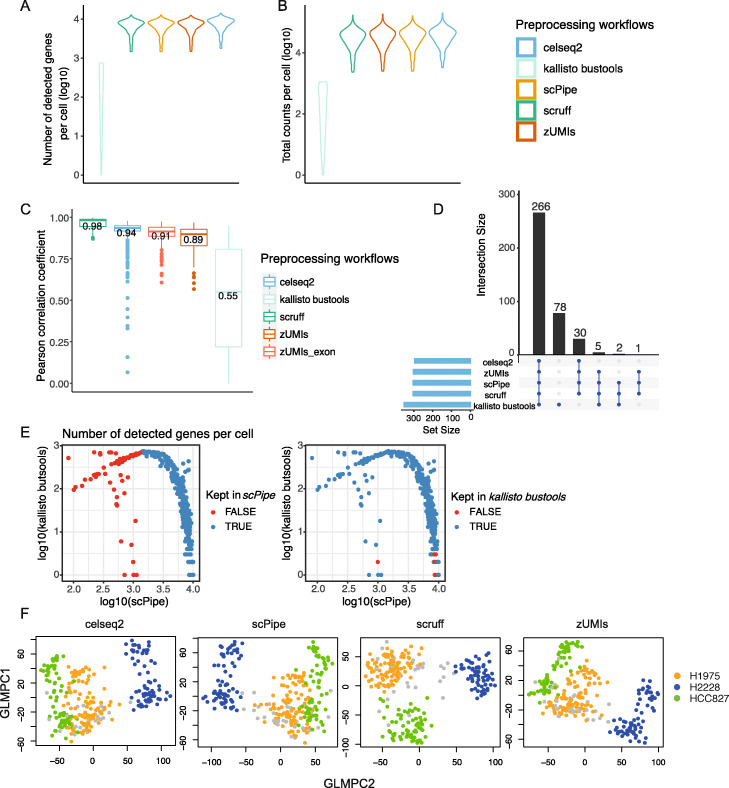
Fig. 3.
Comparing gene expression quantification of different scRNA-seq preprocessing workflows on the plate-based 3 cell line mixture (plate_3cell-line) dataset. A The number of detected genes per cell and B total counts per cell (both on a log10-scale). C The Pearson correlation coefficients between the gene counts of scPipe and other preprocessing workflows. Median values of the correlation coefficients are labelled. D After filtering, an UpSet plot displays the overlap of retained cells across workflows. E The number of detected genes per cell from kallisto bustools and scPipe are plotted in a pairwise manner. Colors represent whether a cell was kept after filtering with scPipe (left panel) and kallisto bustools (right panel). F GLMPCA plots for each preprocessing workflow, with colors representing the different cell lines included in this dataset. Cells that were not common between workflows are colored in grey