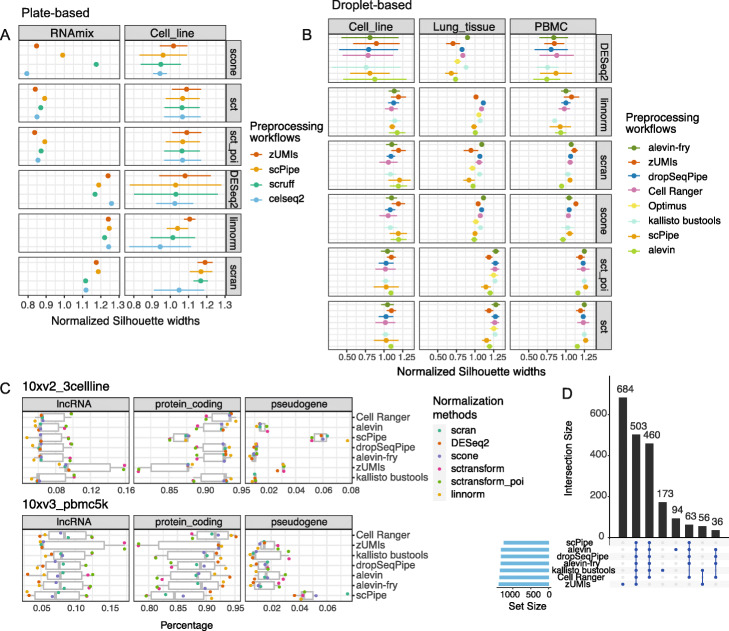
Fig. 6.
Comparing the performance of different scRNA-seq preprocessing workflows and normalization methods. A Dot plots (mean silhouette widths ± s.d) for plate-based datasets and B droplet-based datasets. Colors denote different preprocessing workflows. Silhouette widths are calculated based on known cell labels after applying different normalization methods and normalized against the silhouette widths obtained without any normalization. C The percentage of genes biotypes of lncRNAs, protein coding genes, and pseudogenes among HVGs on the 10xv2_3cell-line and 10xv3_pbmc5k datasets. D An UpSet plot displays the overlap in protein coding genes among the HVG list from different workflows obtained using scran normalized counts for the 10xv3_pbmc5k dataset