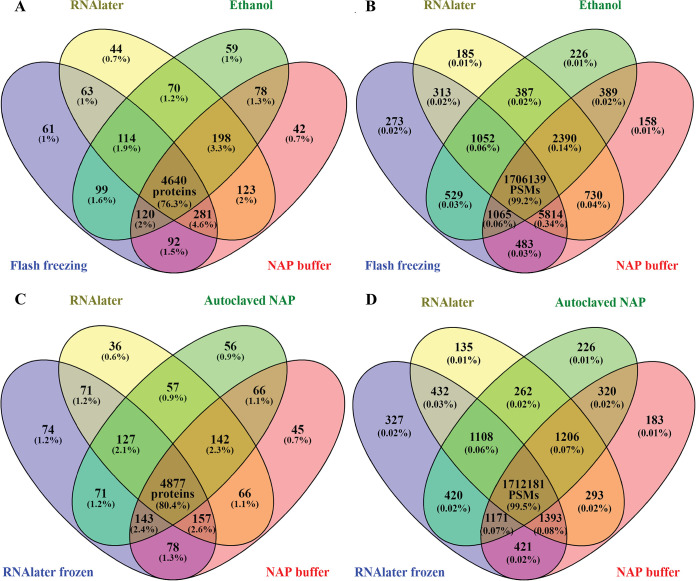
FIG 2.
Over 76% of microbial, host, and dietary protein identifications overlapped between treatments, and these proteins account for more than 99% of all PSMs. Replicates of both time points were combined (n = 8 samples/treatment except for ethanol, for which n = 7). Proteins were included if they were identified with an FDR of <5% and at least one protein unique peptide and were present in at least three samples in the whole data set. The number of PSMs that the proteins represent are displayed in panels B and D. (A and B) Comparison of treatments that differed most in terms of physical/chemical properties: flash-freezing, RNAlater, 95% ethanol, and NAP buffer. (C and D) Comparison of the chemically similar treatments (RNAlater and RNAlater-like treatments): RNAlater, RNAlater frozen, NAP buffer, and autoclaved NAP.