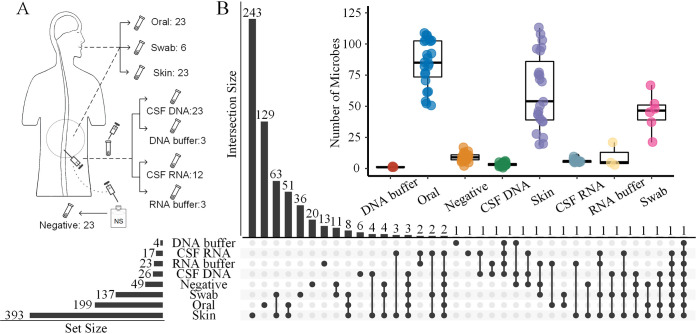
FIG 1.
Study design and numbers of microbes detected in different of types of specimens. (A) Experimental design in this study. CSF and matched control samples (positive controls, oral and skin swab samples; negative controls, saline solution) collected from 23 pregnant women, along with DNA/RNA extraction buffer samples (number indicates the number of specimens) and sterile swabs were sequenced for metagenomic and metatranscriptomic analysis (see Materials and Methods). (B) An overview of microbes detected in each sample type. The number of microbes detected in each sample and species shared between different samples are shown in the UpSet plot, with the dots representing intersections among sample types and the bars representing the number of microbes for each sample type (horizontal bars) and the number shared for each intersection type (vertical bars). The inset shows a box plot summarizing the distributions of the numbers of species detected for different sample types.