Significance
In this study, we show that the inactivation of the gli3 gene in medaka fish results in the formation of larger dorsal and paired fins. These mutant fins display multiple radial bones and fin rays which resemble polydactyly in Gli3-deficient mice. Our molecular and genetic analyses indicate that the size of fish fins is controlled by an ancient mechanism mediated by SHH-GLI signaling that appeared prior to the evolutionary appearance of paired fins. We also show that the key regulatory networks that mediate the expansion of digit progenitor cells in tetrapods were already in place in the fins of the last common ancestor between ray and lobe-finned fishes, suggesting an ancient similarity between distal fins and digits.
Keywords: fin growth, Gli3, Shh, fin-to-limb transition
Abstract
One of the central problems of vertebrate evolution is understanding the relationship among the distal portions of fins and limbs. Lacking comparable morphological markers of these regions in fish and tetrapods, these relationships have remained uncertain for the past century and a half. Here we show that Gli3 functions in controlling the proliferative expansion of distal progenitors are shared among dorsal and paired fins as well as tetrapod limbs. Mutant knockout gli3 fins in medaka (Oryzias latipes) form multiple radials and rays, in a pattern reminiscent of the polydactyly observed in Gli3-null mutant mice. In limbs, Gli3 controls both anterior–posterior patterning and cell proliferation, two processes that can be genetically uncoupled. In situ hybridization, quantification of proliferation markers, and analysis of regulatory regions reveal that in paired and dorsal fins, gli3 plays a main role in controlling proliferation but not in patterning. Moreover, gli3 down-regulation in shh mutant fins rescues fin loss in a manner similar to how Gli3 deficiency restores digits in the limbs of Shh mutant mouse embryos. We hypothesize that the Gli3/Shh gene pathway preceded the origin of paired appendages and was originally involved in modulating cell proliferation. Accordingly, the distal regions of dorsal fins, paired fins, and limbs retain a deep regulatory and functional homology that predates the origin of paired appendages.
A fundamental issue in vertebrate biology is the relationship between fins and limbs. Limbs have a characteristic skeletal pattern of stylopod, zeugopod, and autopod, with a distal region composed of digits and mesopodial bones. While fossil taxa reveal intermediates in these conditions (1–3), the appendages of extant actinopterygians lack common features that allow comparison among the distal regions. Teleosts, for example, have fins with both endochondral and dermal bones. The distal fin typically has a series of cartilage elements with no obvious homology to digits or mesopodials, while rays develop as dermal, not endochondral, bones. Despite these dramatic differences in the distal anatomy of limbs and fins, the terminal region of fins reveals molecular similarities to limbs. Limbs have a characteristic pattern of two phases of expression of Hox genes, an early one that is associated with the specification of the stylopod and zeugopod and a later one associated with the formation of digits. Recent studies of the pattern of expression, function, and cell lineage of Hox-expressing cells in fish reveal that they have late-phase activity that is comparable to that of limbs (4). Lacking, however, is a knowledge of how deep these homologies extend in the tree of life. Are these kinds of similarities common to paired fins or are they a property of diverse paired and unpaired appendages of gnathostomes?
Gli3, a transcription factor expressed from early to late phases of limb development, has been shown to play multiple roles during limb morphogenesis. Gli3 acts to set up the anterior–posterior (AP) axis of the limb bud and restricts Shh pathway activation to the posterior distal margin of appendages (5–8). This activity is mediated by the interactions of Gli3 with the Hand2 and Hoxd transcription factors. Gli3-deficient mice show Shh pathway derepression in the anterior limb bud margin, leading to the anterior expansion of the expression domains of posterior markers and concordant down-regulation of anterior transcription factors (9–14). Morphologically, these mutants reveal polydactylous manus and pes along with soft-tissue fusion of digits. Most importantly, mouse limb buds lacking both Shh and Gli3 have identical gene expression patterns to those of Gli3 single mutants and their limb skeletons are indistinguishable (12, 13). More recent studies have revealed that the roles played by Gli3 in patterning and cellular proliferation during appendage morphogenesis can be genetically uncoupled. The control of the proliferative expansion of the autopod progenitors is hence key to constrain the number of digits to the pentadactyl pattern (15). While Gli3 functions and interactions with Shh are essential features of limb development, little is known about its role in fins. This deficit is unfortunate because an understanding of this issue could reveal the origin of distal patterns between the two organs. To address this, we generated gli3-knockout (KO) mutants in medaka, a teleost fish with a single copy of shh, to assess gene expression, regulation, and ultimately evolution of the morphoregulatory mechanisms mediated by Gli3 in vertebrate appendages.
Results
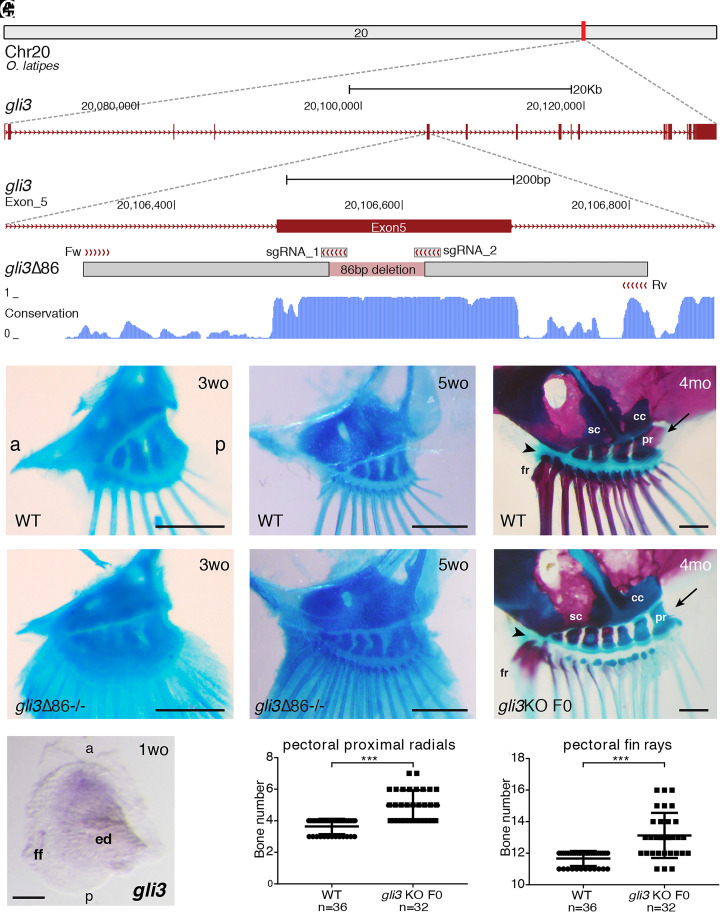
To explore the role of gli3 in medaka fin patterning, we deployed CRISPR-Cas9 to disrupt its coding region via an 86-bp deletion in exon 5 (Fig. 1A and SI Appendix, Fig. S1). This mutation generates a frameshift that truncates the protein in the middle of the repressor domain. Adult fins could not be analyzed, as fish homozygous for this gli3-inactivated allele die between 2 and 5 wk of age, probably reflecting the pleiotropic functions of gli3 in multiple tissues (SI Appendix, Fig. S1). By weeks 3 to 5, however, multiple anomalies were observed in the pectoral fin skeleton of gli3 mutant escapers. In particular, these fins had an expanded number of radials and rays (Fig. 1 B, C, E, and F). Transient gli3 mutants raised to adulthood displayed greatly expanded fins, with multiple supernumerary elements (Fig. 1 D, G, I, and J) in which mineralization appeared delayed (Fig. 1 D and G). Hence, gli3-deficient fish are similar to mouse and human Gli3/GLI3 polydactylous mutants in having expanded appendages with extra bones. This skeletal analysis of pectoral fins also revealed that key morphological AP asymmetries appear unaltered in gli3 mutants in comparison with wild-type (WT) controls (Fig. 1 D and G) (16).
Fig. 1.
Medaka gli3 mutants show an increased number of pectoral fin skeletal elements. (A) Stable lines harboring Δ86 deletions in medaka gli3 exon 5 were generated by CRISPR-Cas9. The diagram shows the position of the deletion relative to the sgRNAs and primers used for screening along with the box of conservation with four fish species (stickleback, fugu, tetraodon, and zebrafish). The Δ86 deletion results in the generation of a premature STOP codon that truncates the predicted protein upstream of the zinc-finger DNA-binding domain of Gli3. (B–G) Alcian blue and Alizarin red staining of pectoral fin skeletons reveals a significantly increased number of proximal radial bones and fin rays in stable (E and F) and transient (G) gli3 mutants, although major anatomical AP asymmetries appear unaltered. Note that, when compared with ones in the anterior margin, the two posterior proximal radials (pr) are larger in both gli3 crispants and WT fish and articulate with the coracoid (cc) bone (arrows in D and G). In the anterior margin of the fin, both the rest of the proximal radials and the anterior-most fin rays (fr) articulate directly with the scapula (sc) in both WT and transient gli3 mutants (arrowheads in D and G). (B and C) n ≥ 14 fins. (E and F) n ≥ 12 fins. (Scale bars, 250 μm.) mo, months-old; wo, weeks-old. (H) WT pectoral fin showing the expression of gli3 in the anterior region of the developing pectoral fin bud (n = 10). (Scale bar, 100 μm.) ed, endochondral disk; ff, fin fold. (I and J) Quantification of skeletal elements in adult (4-mo-old) gli3 crispants. Each point in the graphs represents the measurement of bone number in a single fin (n = 36 WT, n = 32 gli3 KO F0). An unpaired t test was used for the statistical analysis of skeletal element number. ***P = 5.06 × 10−10 for the comparison between WT (mean 3.639) and gli3 crispant (mean 4.969) pectoral proximal radial bones. ***P = 2.33 × 10−7 for the comparison between WT (mean 11.67) and gli3 crispant (mean 13.13) pectoral fin rays. Bone-staining procedures in juvenile (B, C, E, and F) and adult fish (D and G) were performed in three independent experiments. a, anterior; p, posterior.
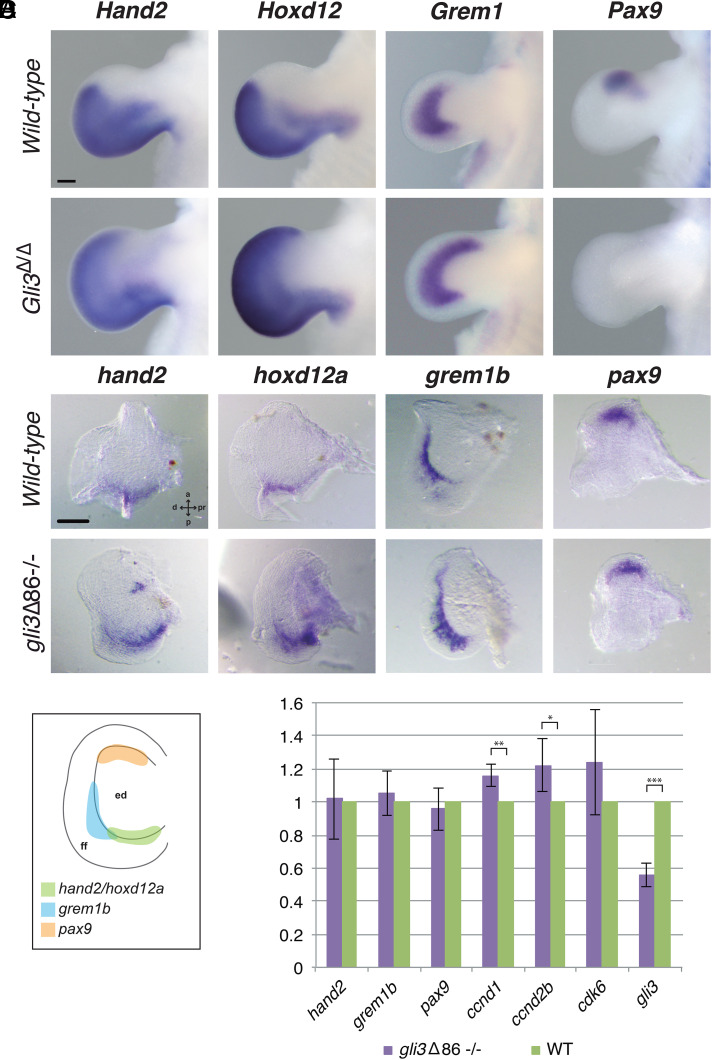
Next, we examined the expression profiles of genes involved in appendage patterning, in particular those known in tetrapods to interact with Gli3 during limb bud outgrowth. Similar to tetrapod limbs, gli3 expression in medaka is higher in the anterior margin of the pectoral fin bud, while shh remains restricted to the posterior-most mesenchyme (Fig. 1H and SI Appendix, Fig. S2). In mouse limb buds, Gli3 inactivation causes the anterior expansion of posterior markers such as Hand2, 5'Hoxd, and Grem1 and down-regulation of anteriorly expressed genes (e.g., Pax9; Fig. 2A) (9, 10, 12–14). In contrast, in situ hybridization (ISH) analysis in medaka gli3 mutant pectoral fins at stage 36 (6 d postfertilization [dpf]) revealed no obvious changes in the expression domains of these genes, as their expression patterns appear largely similar to those of WT embryos (Fig. 2B). At later stages (8 dpf), we noted that, while hoxd12a expression was low in both WT and gli3 mutants, there was an evident expansion of the grem1b expression domain along the rim of the endochondral disk in gli3-deficient pectoral fin buds (SI Appendix, Fig. S3). Because Gli3 has also been shown to control the proliferative expansion of mouse autopod progenitors through the direct transcriptional modulation of cell-cycle regulators (15), we next used qRT-PCR to assess the expression levels of gli3-shh proliferative targets in fin buds (Fig. 2D). This analysis revealed the up-regulation of ccnd1 and ccnd2 in gli3-deficient fins, uncovering a role for Gli3 in controlling distal proliferation during fin outgrowth. Both the deregulation of G1–S cell-cycle modulators and of Grem1 are features observed upon the late, conditional inactivation of Gli3 in mouse handplate progenitors, which develop polydactyly in the absence of AP polarization defects (15).
Fig. 2.
Expression of gli3 target genes in WT and gli3-deficient pectoral fin buds. (A) Hand2, Hoxd12, and Grem1 expression is anteriorly expanded and Pax9 expression is lost in Gli3-deficient E11.5 mouse limb buds (n = 3 per marker and genotype). (Scale bar, 200 µm.) (B) ISH assays in WT and gli3Δ86−/− medaka pectoral fins. At 6 dpf (stage 36), the inactivation of gli3 does not greatly affect the expression pattern of hand2, hoxd12a, grem1b, or pax9, all genes involved in limb patterning (n ≥ 4 per marker and genotype). d, distal; pr, proximal. (Scale bar, 100 µm.) (C) Schematic representation of the hand2, hoxd12a, grem1b, and pax9 expression domains in 6-dpf WT medaka pectoral fins. Note that the grem1b expression domain (shown in B) runs along the margin of the endochondral disk (ed) and extends into the fin fold. (D) Gene expression quantification by qPCR in WT and gli3Δ86−/− medaka pectoral fins at 11 dpf. The relative expression of the proliferation regulators ccnd1 and ccnd2b is significantly increased in mutant fins. Mutant values (purple bars) are normalized against WT values (green bars), and represented as mean ± SD. Note that the gli3Δ86 allele is still transcribed in homozygous mutant fins, although at lower levels due to nonsense-mediated mRNA decay. nhand2 = 4, ngrem1b = 3, npax9 = 3, nccnd1 = 2, nccnd2b = 4, ncdk6 = 3, ngli3 = 4; ***Pgli3 = 9.026 × 10−9, **Pccnd1 = 0.0022, *P ≤ 0.05, Pccnd2b = 0.011.
Our morphological, gene expression, and qPCR analyses point toward a role for gli3 in cell proliferation, rather than AP patterning, during pectoral fin development. To explore this hypothesis and reveal its phylogenetic distribution, we analyzed Gli3 chromatin immunoprecipitation sequencing (ChIP-seq) data from mouse limb buds. This strategy revealed that Gli3 preferentially binds at the promoters of these proliferation genes (SI Appendix, Fig. S4). These regulatory regions are present in diverse tetrapods and fishes, revealing their phylogenetic generality. In contrast, Gli3 regulation of patterning genes (e.g., Grem1, Hand2, Pax9, and Hoxd genes) takes place mainly through distal cis-regulatory elements (CREs) that are mostly not conserved in fish (17, 18). In particular, most of the Gli3-binding CREs previously reported within the regulatory landscapes of Hand2 and Hoxd genes appear to be tetrapod innovations (SI Appendix, Figs. S4 and S5) while, of the two characterized Grem1 enhancers bound by Gli3, only one [GRS1 (18)] is deeply conserved in all vertebrates, and could mediate the late AP expansion of grem1 in medaka gli3 mutant fins (SI Appendix, Fig. S4). Overall, these data support the notion that the incorporation of Gli3 in early patterning events in appendages is a more recent novelty within derived gnathostomes that evolved through the progressive acquisition of new distal CREs.
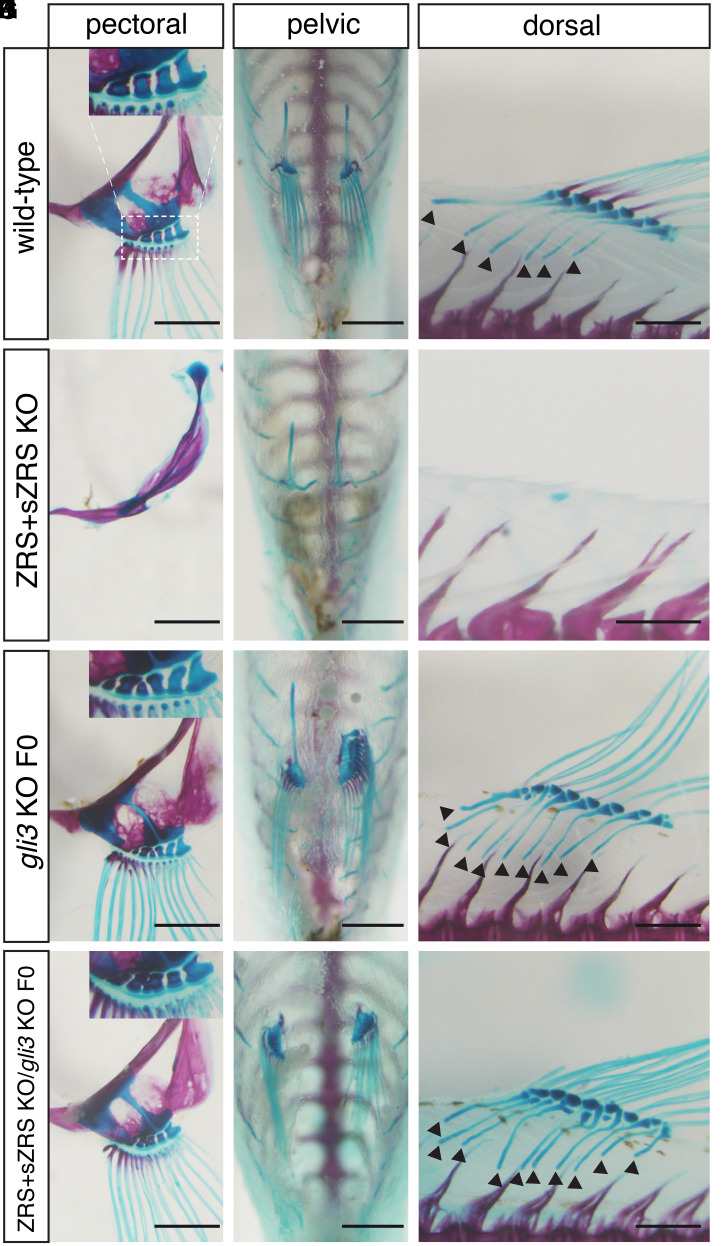
A main function of Shh signaling is to antagonize the constitutive proteolytic processing of Gli3 to its transcriptional repressor isoform (19). Genetic analysis in mice has shown that Gli3 deficiency is able to rescue the loss of distal limb bones observed in Shh-null embryos, leading to polydactyly that is identical to that observed upon Gli3 inactivation alone (12, 13). To examine this relationship in medaka, we produced gli3/shh double mutants transiently, as gli3-deficient fish are not viable. Transient inactivation of gli3 in a ZRS+sZRS shh mutant background (20) is sufficient to rescue the shh loss-of-function phenotype (agenesis of pectoral, pelvic, and dorsal fins). Analogous to the genetic interaction observed in mutant mouse autopods, the fin skeleton of ZRS+sZRS/gli3 F0 double mutants resembles those of gli3 crispants. This effect was seen in both paired and unpaired fins, as supernumerary bones were seen in pectoral, pelvic, and dorsal fins (Fig. 3 and SI Appendix, Fig. S6). Interestingly, as observed previously in the ZRS+sZRS shh mutant (20), the anal fin is not affected by any of these mutant conditions (SI Appendix, Fig. S7).
Fig. 3.
Mosaic inactivation of gli3 in fins lacking shh completely rescues the formation of dorsal, pelvic, and pectoral fins. Skeletal staining and fin morphology in adult fish. (A–F) In contrast to WT (A–C), all fin elements are absent in the dorsal, pelvic, and pectoral fins from ZRS+sZRS mutant fish (D–F). (G–I) CRISPR-Cas9 disruption of gli3 significantly increases the number of pectoral and dorsal fin bone elements (black arrowheads in I) compared with WT (A–C). (J–L) gli3 down-regulation in the homozygous ZRS+sZRS shh mutant background totally rescues the dorsal, pelvic, and pectoral fin phenotypes. Note that there is a delay in the ossification of dorsal fin rays in gli3 single and gli3/ZRS+sZRS compound mutants. (Scale bars, 1 mm.) Microinjection of the gli3 CRISPR mixture and bone-staining procedures were performed in three independent experiments.
Discussion
Altogether, our results show that the presence of the shh/gli3 regulatory network in fish fins, so vital for limb formation and digit patterning, is primitive to limbs. Moreover, its functions in unpaired dorsal fins, widely recognized precursors of paired appendages, suggest that the recruitment of this network may have preceded the origin of paired fins themselves.
The correlation of expanded radials and rays in gli3 fin mutants with the polydactyly in mouse Gli3 mutants points to a deep homology among the distal tissues of gnathostome appendages. Our analyses suggest that the primitive function of the Shh/Gli3 module in appendages was to control the proliferative expansion of the distal mesenchyme. Interestingly, the transcriptional control of cell-cycle effectors is an ancient feature of HH signaling, as Ci, the fly ortholog of Gli3, also directly regulates several cyclin genes (21, 22). In contrast, the fully wired AP patterning systems controlled by Gli3 likely evolved later during evolution, probably in the tetrapod lineage through the appearance of novel far-acting cis-regulatory regions (23, 24). As expected, given their dependence on SHH signaling for their posterior up-regulation (25–28), some deeply conserved Gli-binding CREs are present in the Hoxd and Grem1 genomic landscapes. Moreover, our observation that the grem1b expression domain is expanded in gli3-deficient fin buds at 8 dpf suggests that some aspects of grem1b regulation are controlled by Gli3 at these advanced stages. This is highly reminiscent of the situation in the mouse, as failure to terminate Grem1 expression in the anterior margin of the handplate leads to a delay in chondrogenic differentiation that contributes to the polydactyly observed in Gli3-deficient limb buds (15). The highly conserved GRS1 enhancer is a good candidate to implement some aspects of Gli3-mediated repression of Grem1/grem1b at these late stages of appendage development, as its activity is sensitive to Gli3 gene dosage in the mouse (18). Finally, another possibility explaining the lack of apparent polarization defects in medaka gli3 mutants would be that SHH-mediated AP patterning in early teleost fins (25, 26) is mediated in a gli3-independent manner by a Gli2 factor(s), capable of being processed into GLI activator and repressor isoforms (29, 30). Interestingly, Gli2 has been genetically shown to cooperate with Gli3 in providing posterior identity to the mouse autopod (31).
Overall, our results imply that the distal regions of appendages have a common evolutionary origin and that the Shh/Gli3 network was modified in fish and tetrapod lineages to produce fin radials and rays in the former and digits in the latter. Interestingly, the only appendage that does not follow these rules, the anal fin, also has anomalous patterns of shh regulation. The absence of these networks in anal fins points to a separate evolutionary origin for anal fins, presumably by independent cooption of fin patterning networks in a novel site.
Materials and Methods
Animal Experimentation.
All experiments involving fish and mice performed in this work conform to European Community standards for the use of animals in experimentation and were approved by the ethical committees from the Universidad Pablo de Olavide, Universidad Mayor, and Consejo Superior de Investigaciones Científicas and the Andalusian government.
Fish Stocks.
Medaka WT (iCab) and ZRS+sZRS KO [Δ3.4kb (20)] strains were maintained and bred under standard conditions (32). Embryos were staged in hours postfertilization as previously described (33). Medaka gli3_Δ86 and ZRS_Δ3.4kbs mutant alleles were maintained in heterozygosis due to the higher lethality of the homozygous mutants. gli3_Δ86 homozygous null larvae died between 2 and 5 wk of age.
Skeletal Staining.
Alcian blue and Alizarin red staining experiments were performed as previously described (20, 34). In brief, fish were fixed with 10% neutral-buffered formalin overnight or longer. After several washes with deionized water, cartilage was stained overnight using a 0.1% Alcian blue solution (Alcian blue 8GX; PanReac AppliChem) in 30% acetic acid and 70% ethanol. Fish were then washed with deionized water and changed to a solution containing 1% trypsin from bovine pancreas (PanReac AppliChem) and 30% saturated sodium borate for 8 h (or longer) with gentle shaking at room temperature. After trypsin enzyme treatment, specimens were rinsed several times with deionized water and transferred to 0.5% aqueous KOH solution. Fish bones were finally stained overnight with a solution containing saturated Alizarin red S (PanReac AppliChem) in 0.5% KOH. After several washes with a 0.5% KOH solution, fish were gradually transferred to glycerol for documentation. Specimens were visualized with an Olympus SZX16 binocular microscope and photographed with an Olympus DP71 camera.
CRISPR-Cas9 Design and gli3 Mutant Generation.
Two single guide RNAs (sgRNAs) targeting exon 5 of medaka gli3 were designed using the CRISPRscan (35) and CCTop (36) CRISPR design online tools. sgRNAs were generated and purified for injection as previously described (37). For sgRNA generation, the following primers were aligned (by PCR) to a universal CRISPR primer: gli3 exon 5 sgRNA1: 5'-taatacgactcactataGGGCGGATGTAGTCCATGTAgttttagagctagaa-3' and gli3 exon 5 sgRNA2: 5'-taatacgactcactataGGGGTGAGATCCGAATGAGGgttttagagctagaa-3' (in both primers the target site is identified by capital letters). Following synthesis, 5 nL of a solution containing both sgRNAs at a concentration of 40 ng/μL and Cas9 protein (Addgene; 47327) at a concentration of 250 ng/μL (38) were injected into one-cell-stage medaka embryos. Oligos used for screening of genomic DNA deletions flanking CRISPR target sites were the following: forward primer 5'-CGTGAGTTTCACAGCAACAATTA-3' and reverse primer 5'-CAGCCTCACTGATCAATTTCAG-3'. Mutations in gli3 were analyzed by standard PCR and gel electrophoresis as the distance between both sgRNA protospacer adjacent motif (PAM) sequences was 82 bp, long enough to create a deletion easily detected by a shift in the PCR band. Specific deletions in the gli3 gene were further analyzed by Sanger sequencing of the PCR product from F1 embryos (Stab Vida).
Statistical Analyses.
The number of pectoral fin proximal radial bones, pectoral fin rays, and dorsal proximal pterygiophores was manually counted in adult WT and gli3 crispant fish after a bone-staining procedure. Differences in the number of skeletal elements between both groups were tested by applying an unpaired t test using GraphPad Prism software. Paired two-tailed t tests were used for the statistical analysis of average differences in gene expression levels between mutant and WT samples as measured by qRT-PCR.
Medaka In Situ Hybridization.
Depending on the genomic location of the designed primers, antisense digoxigenin-labeled RNA probes were prepared from 4-dpf medaka complementary DNA (cDNA) or gDNA (SI Appendix, Table S1). shh (20), hand2, hoxd12a, grem1b, and pax9 RNA probes were synthesized by cloning the DNA-amplified region using the StrataClone PCR Cloning Kit (240205-5; Agilent Technologies; hoxd12a) or pGEM-T Easy Vector (A1360; Promega; pax9, grem1b, and hand2), and these linearized vectors were used as templates for RNA transcription. The gli3 probe was directly transcribed from the amplified DNA since the SP6-RNA polymerase promoter sequence (labeled red in SI Appendix, Table S1) was included in the primers used for amplicon amplification.
Heterozygous animals were mated in order to collect embryos to perform ISH assays. The embryos were maintained at 28 °C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.4 mM CaCl2, 0.16 mM MgSO4, and 0.00003% methylene blue) until 4, 6, 8, and 11 dpf and fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS) for 48 h at 4 °C. Embryos were manually dechorionated when needed (stages 4 to 8 dpf), dehydrated through an increasing MeOH washing series, and kept at −20 °C until the experiments were performed. Overall, the specimens were prepared, hybridized (2 ng probe per microliter), and stained as previously described (39, 40). Embryos at stages older than 4 dpf were permeabilized with 10 µg of ProtK for 7 min.
In order to analyze the expression of the different genes in mutant and WT siblings, samples were genotyped after ISH assays. gDNA was extracted using Chelex 100 sodium form (C7901; Sigma) from a piece of tail dissected from each individual larva and standard PCR reactions were performed with primers flanking the deleted region (Fig. 1 and SI Appendix, Table S1). Subsequently, gene expression patterns in homozygous null and WT pectoral fins were analyzed and documented. Both pectoral fins from each genotyped larva were dissected and transferred to a drop of 3% methyl cellulose (M0387; Sigma-Aldrich) on a slide for documentation using an Olympus SZX16 (model SZX2-ILLB) binocular scope and an Olympus DP71 camera.
Quantitation of Transcript Levels in Medaka Fins by qRT-PCR.
Larvae were raised as described above until 11 dpf and deeply anesthetized with 160 mg/L of tricaine (ethyl 3-aminobenzoate methanesulfonate salt; MS-222; Merck) before dissecting their pectoral fins. The dissection was performed in a drop of embryos’ medium and the fins were rapidly moved to an ice-cold drop of PBS. Every pair of dissected pectoral fins coming from a single larva was transferred to a separate 1.5-mL tube containing 50 µL TRIsure (BIO-38032; Bioline) and stored at −20 °C until larvae were genotyped (Medaka In Situ Hybridization). In order to obtain sufficient RNA material, each biological replicate consisted of 20 pairs of pectoral fin buds per genotype (WT and gli3-deficient). RNA was extracted following the TRIsure manufacturer’s instructions and equivalent amounts of mutant and WT RNA were used to synthesize cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems–Thermo Fisher Scientific; 4368814). The expression levels of medaka hand2, grem1b, pax9, ccnd1, ccnd2b, cdk6, and gli3 in the developing fins were quantified through qRT-PCR (CFX96 Real-Time C1000 thermal cycler; Bio-Rad) and normalized to the expression level of the housekeeping gene ef1a (SI Appendix, Table S1). qPCR reactions were performed in triplicate from two to four biological replicates using iTaq Universal SYBR Green Supermix (Bio-Rad; 172-5124; nhand2 = 4, ngrem1b = 3, npax9 = 3, nccnd1 = 2, nccnd2b = 4, ncdk6 = 3, ngli3 = 4). The expression levels in mutant samples were calculated in relation to WT controls (average set to 100%). Assuming a normal distribution of the data, a paired two-tailed t test was performed to test the significance of differences among sample averages. As the gli3Δ86 mutation induces a frameshift that results in premature stop codons, gli3 transcription levels are strongly reduced in gli3Δ86 homozygous fins due to nonsense-mediated messenger RNA (mRNA) decay.
Mouse Experiments.
Embryonic day 11.5 (E11.5) (44 to 48 somites) Gli3-deficient and WT embryos were processed for ISH with riboprobes recognizing Hand2, Hoxd12, Grem1, and Pax9 transcripts (n = 3 per marker and genotype), as previously described (15). Gli3 ChIP-seq and assay for transposase-accessible chromatin with sequencing (ATAC-seq) data generated from E10.5 mouse limb buds have been previously published (41, 42).
Acknowledgments
This work would have been impossible without the contribution of José Luis Gómez-Skarmeta, a beloved friend and mentor whose passion for science was only surpassed by his generosity and kindness. We thank members of the J.L.G.-S. laboratory for fruitful discussions. J.L.G.-S. received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (Grant Agreement 740041) and the Spanish Ministerio de Economía y Competitividad (Grant BFU2016-74961-P) and was also supported, together with J.L.-R. and J.R.M.-M., by institutional grant Unidad de Excelencia María de Maeztu (MDM-2016-0687, to the Department of Gene Regulation and Morphogenesis of the Centro Andaluz de Biología del Desarrollo). J.L.-R. and J.R.M.-M. are supported by the Spanish Ministerio de Ciencia e Innovación (Grants BFU2017-82974-P/PID2020-113497GB-I00 and BFU2017-86339-P/PID2020-112566GB-I00, respectively). N.S. is supported by grants from the Brinson Foundation. J.L. is financed by the Chilean FONDECYT agency (Grant 11180727).
Footnotes
Reviewers: M.R., Universidad de Cantabria; and J.M.W., Universitat Konstanz.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100575118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
Previously published data were used for this work (PMID: 31989924; PMID: 32268095).
Note Added in Proof. During the review of this manuscript, another study (43) reported an additional highly-conserved Grem1 enhancer, termed CRM5, that is, as GRS1 (renamed as CRM2), also regulated by Gli3.
References
- 1.Shubin N. H., Daeschler E. B., F. A. Jenkins, Jr, The pectoral fin of Tiktaalik roseae and the origin of the tetrapod limb. Nature 440, 764–771 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Boisvert C. A., Mark-Kurik E., Ahlberg P. E., The pectoral fin of Panderichthys and the origin of digits. Nature 456, 636–638 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Cloutier R., et al. , Elpistostege and the origin of the vertebrate hand. Nature 579, 549–554 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Nakamura T., Gehrke A. R., Lemberg J., Szymaszek J., Shubin N. H., Digits and fin rays share common developmental histories. Nature 537, 225–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galli A., et al. , Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 6, e1000901 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.te Welscher P., Fernandez-Teran M., Ros M. A., Zeller R., Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 16, 421–426 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakany J., Zacchetti G., Duboule D., Interactions between HOXD and Gli3 genes control the limb apical ectodermal ridge via Fgf10. Dev. Biol. 306, 883–893 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Osterwalder M., et al. , HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev. Cell 31, 345–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büscher D., Bosse B., Heymer J., Rüther U., Evidence for genetic control of sonic hedgehog by Gli3 in mouse limb development. Mech. Dev. 62, 175–182 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Zúñiga A., Zeller R., Gli3 (Xt) and formin (ld) participate in the positioning of the polarising region and control of posterior limb-bud identity. Development 126, 13–21 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Hill P., Götz K., Rüther U., A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Dev. Biol. 328, 506–516 (2009). [DOI] [PubMed] [Google Scholar]
- 12.te Welscher P., et al. , Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science 298, 827–830 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Litingtung Y., Dahn R. D., Li Y., Fallon J. F., Chiang C., Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418, 979–983 (2002). [DOI] [PubMed] [Google Scholar]
- 14.McGlinn E., et al. , Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech. Dev. 122, 1218–1233 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Rios J., et al. , GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev. Cell 22, 837–848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwamatsu T., Growth of the medaka (II)—Formation of fins and fin appendages. Bull. Aichi Univ. Educ. 62, 53–60 (2013). [Google Scholar]
- 17.Vokes S. A., Ji H., Wong W. H., McMahon A. P., A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuniga A., et al. , Conserved cis-regulatory regions in a large genomic landscape control SHH and BMP-regulated Gremlin1 expression in mouse limb buds. BMC Dev. Biol. 12, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B., Fallon J. F., Beachy P. A., Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423–434 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Letelier J., et al. , A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet. 50, 504–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duman-Scheel M., Li X., Orlov I., Noll M., Patel N. H., Genetic separation of the neural and cuticular patterning functions of gooseberry. Development 124, 2855–2865 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Biehs B., Kechris K., Liu S., Kornberg T. B., Hedgehog targets in the Drosophila embryo and the mechanisms that generate tissue-specific outputs of hedgehog signaling. Development 137, 3887–3898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheth R., et al. , Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science 338, 1476–1480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onimaru K., et al. , A shift in anterior-posterior positional information underlies the fin-to-limb evolution. eLife 4, e07048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn D., Ho R. K., Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: Implications for the evolution of vertebrate paired appendages. Dev. Biol. 322, 220–233 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Neumann C. J., Grandel H., Gaffield W., Schulte-Merker S., Nüsslein-Volhard C., Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development 126, 4817–4826 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Zúñiga A., Haramis A. P., McMahon A. P., Zeller R., Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature 401, 598–602 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Chiang C., et al. , Manifestation of the limb prepattern: Limb development in the absence of sonic hedgehog function. Dev. Biol. 236, 421–435 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Karlstrom R. O., et al. , Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development 130, 1549–1564 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Ben J., Elworthy S., Ng A. S., van Eeden F., Ingham P. W., Targeted mutation of the talpid3 gene in zebrafish reveals its conserved requirement for ciliogenesis and hedgehog signalling across the vertebrates. Development 138, 4969–4978 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowers M., et al. , Limb anterior-posterior polarity integrates activator and repressor functions of GLI2 as well as GLI3. Dev. Biol. 370, 110–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vázquez-Marín J., et al. , yap1b, a divergent Yap/Taz family member, cooperates with yap1 in survival and morphogenesis via common transcriptional targets. Development 146, dev173286 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Iwamatsu T., Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 121, 605–618 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Bird N. C., Mabee P. M., Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev. Dyn. 228, 337–357 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Moreno-Mateos M. A., et al. , CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J. L., CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10, e0124633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vejnar C. E., Moreno-Mateos M. A., Cifuentes D., Bazzini A. A., Giraldez A. J., Optimized CRISPR-Cas9 system for genome editing in zebrafish. Cold Spring Harb. Protoc. 2016, pdb-prot086850 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Gagnon J. A., et al. , Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9, e98186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jowett T., Lettice L., Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 10, 73–74 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Tena J. J., et al. , Odd-skipped genes encode repressors that control kidney development. Dev. Biol. 301, 518–531 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Lex R. K., et al. , GLI transcriptional repression regulates tissue-specific enhancer activity in response to hedgehog signaling. eLife 9, e50670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tissières V., et al. , Gene regulatory and expression differences between mouse and pig limb buds provide insights into the evolutionary emergence of artiodactyl traits. Cell Rep. 31, 107490 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malkmus J.et al. , Spatial regulation by multiple Gremlin1 enhancers provides digit development with cis-regulatory robustness and evolutionary plasticity. Nat. Commun. 12, 5557 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or SI Appendix.
Previously published data were used for this work (PMID: 31989924; PMID: 32268095).
Note Added in Proof. During the review of this manuscript, another study (43) reported an additional highly-conserved Grem1 enhancer, termed CRM5, that is, as GRS1 (renamed as CRM2), also regulated by Gli3.