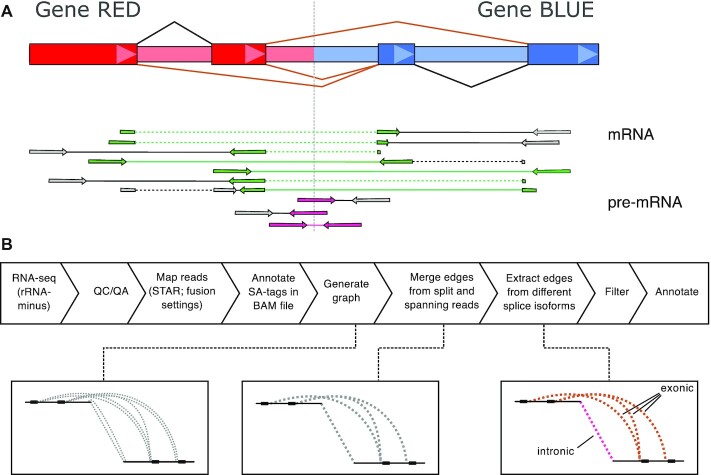
Figure 1:
Overview Dr. Disco pipeline and principle. (A) Schematic representation of fusion-gene RED-BLUE. Owing to relatively large intron sizes, in-gene genomic breakpoints occur most often intronically. The fusion results in different fusion splice isoforms (brown splice junction annotation in fusion gene structure). Fusion splice junction spanning reads form the classical evidence for mature mRNA fusion events (reads marked in green). In rRNA-minus data, intronic pre-mRNA reads (reads marked in pink) may cover causal genomic breakpoints. (B) Pipeline flow chart: RNA-seq data is aligned. Discordant reads are transformed into edges and inserted into a breakpoint graph. In the graph, intronic or exonic derived edges are kept separate. Detection of junctions is performed by analysing the graph for clusters. An additional splice variant correction is applied. Identified junctions are filtered, annotated, and marked intronic or exonic. QC/QA: quality control/quality assurance.