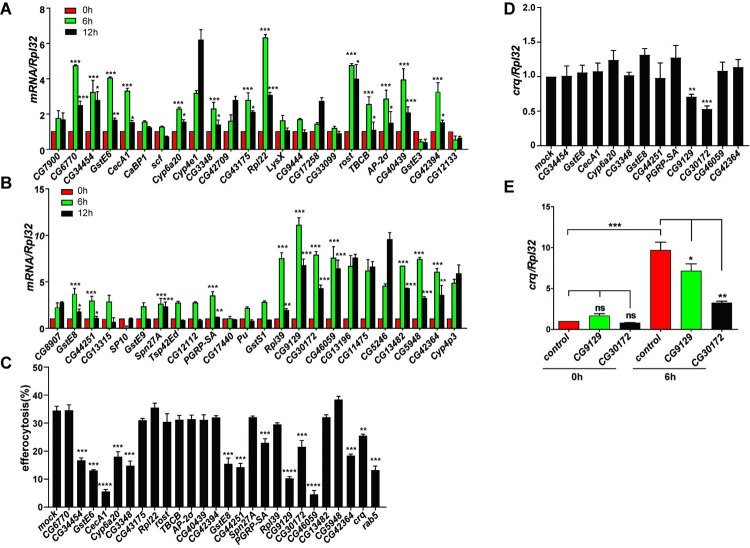
Fig 2. CG9129 is involved in efferocytosis and regulates crq expression in vitro.
A and B. The transcription levels of differentially expressed genes determined by the transcriptomic analysis were validated by qPCR in S2 cells incubated with ACs for 0, 6, and 12 h. Twenty-four genes showed expression patterns similar to that of crq. The data were collected from three independent experiments; 0 h S2 cells were used as the control. The asterisks above the column of the 6 h time-point denote a significantly higher gene expression than that detected at 0 h. The asterisks above the column of the 12 h time-point denote a significantly lower gene expression than that detected at 6 h. C. Efferocytosis efficiency of RNAi-treated S2 cells after 6 h co-incubation with ACs. Each of the identified 24 genes was knocked down individually. The data are representative of three independent experiments. Significant differences were observed in comparison with mock RNAi-treated S2 cells (negative control), or crq and Rab5 RNAi-treated S2 cells (positive controls). D. The crq mRNA levels were quantified by qPCR in RNAi-treated S2 cells showing decreased efferocytosis during 6 h incubation with ACs. The data are relative to the expression determined in mock RNAi-treated S2 cells. E. The relative crq mRNA levels were also quantified by qPCR in control, CG9129 knockdown, and CG30172 knockdown S2 cells in the absence (0 h) or presence of ACs (6 h incubation). Statistical analysis was performed using the two-way ANOVA (A-B, E) or one-way ANOVA (C-D).