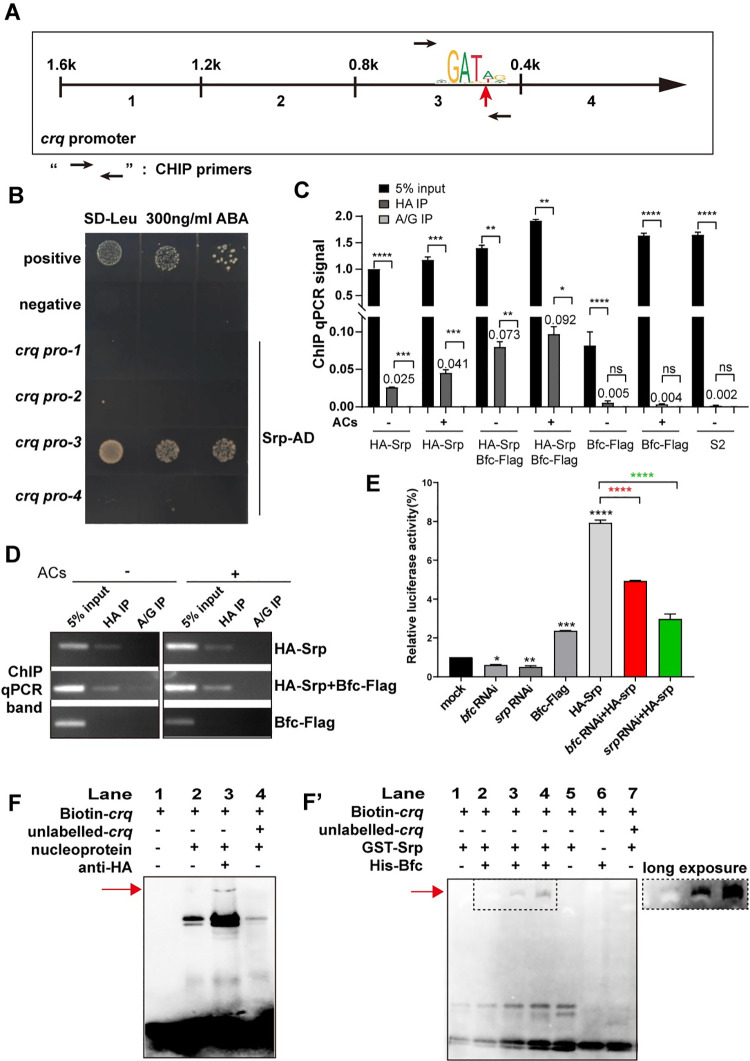
Fig 6. Srp binds to the GATA motif in the crq promoter.
A. Schematic representation of the crq promoter region; the red arrowhead indicates the predicted Srp binding site, and the black arrows indicate the primers used for the ChIP-qPCR. B. Yeast One-hybrid assays with yeast strains containing different prey (Srp-AD)—bait (crq promoter plasmids) combinations. C. The crosslinked chromatin of S2 cells expressing HA-Srp, HA-Srp+Bfc-Flag, or Bfc-Flag, incubated or not with ACs was immunoprecipitated with either anti-HA beads or control protein A/G. The ChIP-qPCR efficiency (compared to 5% input) is shown. Protein A/G was used as the negative control. D. The gel image shows the input DNA or immunoprecipitated DNA bands of the ChIP-qPCR in (C). E. Effect of bfc and srp RNAi treatments on the luciferase activity of crq promoter reporter constructs. All the samples were tested after the addition of ACs. The mean activity with the standard deviation of three independent transfections is represented. Statistical significance was assessed using the one-way ANOVA. F. Nuclear proteins extracted from HA-Srp transfected S2 cells binding to biotin-labeled crq promoter probe and EMSA supershift with and without anti-HA antibodies. The red arrows point to supershifted bands. F’. Purified GST-Srp protein binding to biotin-labeled crq probe and supershift with purified Bfc-6XHis.