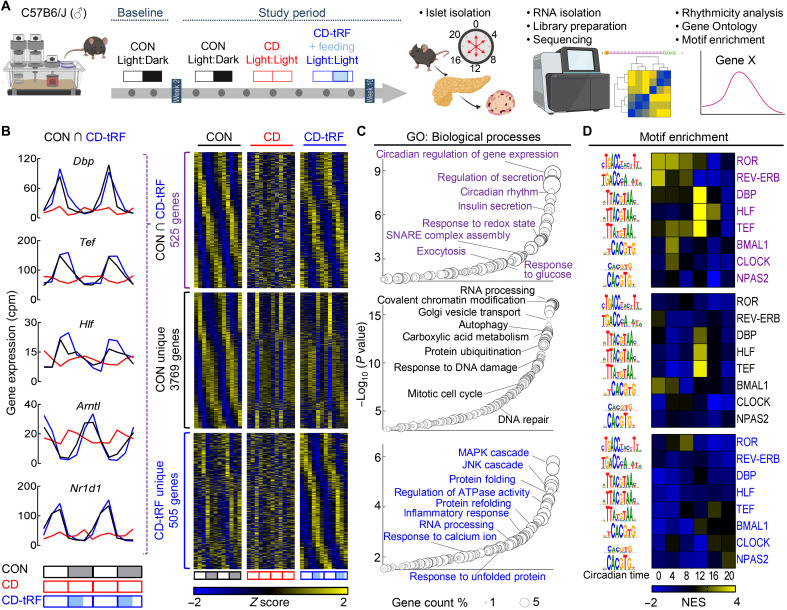
Fig. 3. tRF preserves circadian regulation of gene expression in islets under conditions of global circadian disruption in mice.
(A) Overview of study design. Islets were isolated for RNA-seq from CON, CD, and CD-tRF mice at 4-hour intervals for two independent replicates in vivo. Rhythmic circadian gene expression was determined using MetaCycle (30) with GO and motif enrichment assessed using WebGestalt and i-Cis Target, respectively. (B) Heatmaps representing genes commonly cycling (FDR ≤ 0.1) in CON and CD-tRF islets (top), uniquely cycling in CON islets (middle), and uniquely cycling in CD-tRF islets (bottom). Gene expression was normalized by row Z score. Each column depicts one time point, sampled every 4 hours, starting at CT0 for two independent replicates (n = 12 independent samples per group from n = 2 mice per time point/per group). Inset shows circadian gene expression profiles for Dbp, Tef, Hlf, and Arntl encoding for BMAL1 and Nr1d1 encoding for REV-ERBα commonly cycling in CON and CD-tRF but arrhythmic under CD conditions. (C) Enriched GO: Biological Processes annotated from genes commonly cycling in CON and CD-tRF islets (top), uniquely cycling in CON islets (middle), and uniquely cycling in CD-tRF islets (bottom). Key pathways associated with β cell function are highlighted. (D) Heatmaps representing predicted i-Cis Target enrichment of circadian regulatory motifs (colored by Z-normalized enrichment score) in cis-regulatory regions of rhythmic genes with a peak phase centering around 4-hour intervals starting from CT0. Motif enrichment analysis was assessed in genes commonly cycling in CON and CD-tRF islets (top), uniquely cycling in CON islets (middle), and uniquely cycling in CD-tRF islets (bottom). Highly enriched motifs within cis-regulatory regions of cycling genes are colored yellow.