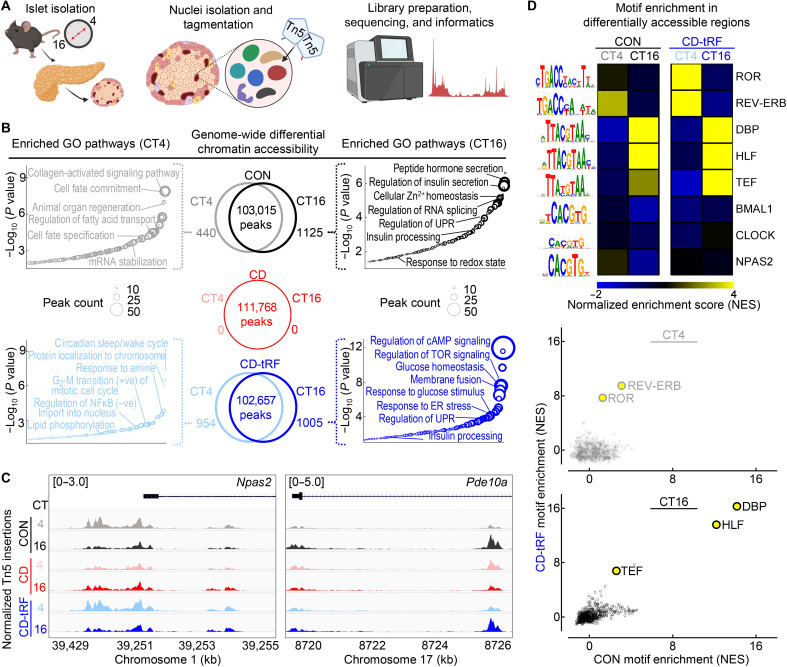
Fig. 4. tRF maintains diurnal regulation of chromatin accessibility in islets under conditions of global circadian disruption in mice.
(A) Overview of study design. Islet cell nuclei were isolated from CON, CD, and CD-tRF mice at CT4 and CT16 time points and subjected to assay for transposase-accessible chromatin using sequencing (ATAC-seq). (B) Venn diagrams depict diurnal (CT4 versus CT16) differentially accessible chromatin regions (FC > 1.5; FDR < 0.1 using Benjamini-Hochberg correction) from ATAC-seq performed in CON (black), CD (red), and CD-tRF (blue) isolated islets (n = 2 independent samples per time point/group from n = 2 mice per time point/group) (center). Enriched GO: Biological Process pathways annotated from differentially accessible chromatin regions at CT4 and CT16 in CON (left) and CD-tRF (right). Key pathways associated with β cell function are highlighted. (C) Integrative Genomics Viewer browser tracks of representative open chromatin region at the Npas2 (left) and Pde10a (right) transcriptional start site CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) samples for CT4 and CT16. Signal is normalized to counts per million reads (cpm) from 0 to 5 cpm (Pde10a) and 0 to 3 cpm (Npas2). Shaded regions represent areas of accessible chromatin. (D) Heatmap representing predicted i-Cis Target enrichment of circadian regulatory motifs (colored by Z-normalized enrichment score) in differentially accessible chromatin regions at CT4 and CT16 time points from CON and CD-tRF islets. Highly enriched motifs in differentially accessible chromatin regions are colored yellow (top). Correlation comparing normalized motif enrichment score from CON and CD-tRF islets isolated at CT4 (middle) and CT16 (bottom). Highly correlated motifs common to CON and CD-tRF are denoted in yellow.