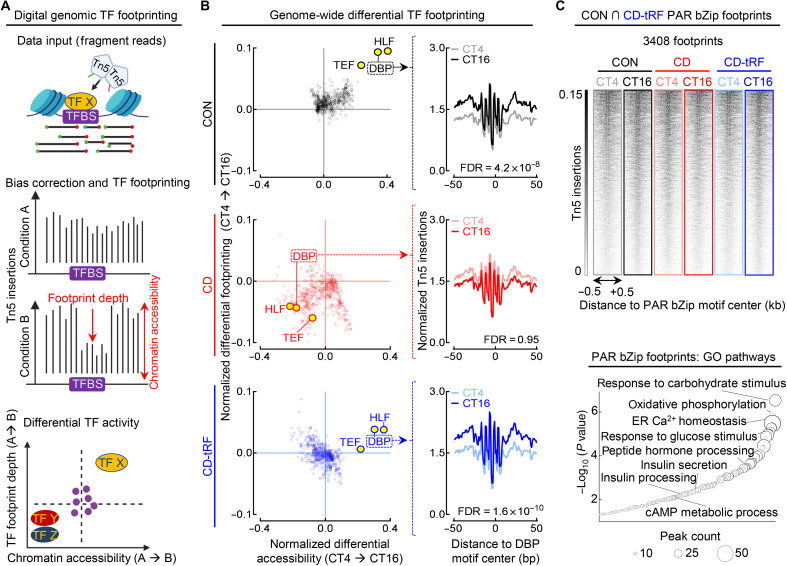
Fig. 5. Digital genomic footprinting reveals that tRF protects diurnal PAR bZip transcription factor activity in islets under conditions of circadian disruption.
(A) Overview of transcription factor (TF) footprinting. Regulatory TFs protect against tagmentation, resulting in a signal decrease (footprint), and allow for identification of bound TFs. Differential changes in footprint depth or accessibility are associated with increased/decreased TF activity between conditions. (B) Global changes in TF footprint depth from CT4 to CT16 representing diurnal differential TF activity (y axis) and flanking accessibility from CT4 to CT16 representing diurnal differential global chromatin accessibility around each TF motif tested (x axis) in CON (black), CD (red), and CD-tRF (blue) islets. PAR bZip (DBP, HLF, and TEF) TFs are highlighted in yellow (left). Mean diurnal DBP footprints in CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) islets at CT4 and CT16. Signal is normalized to the total read depth of each sample and is plotted ±50 bp from DBP motif center (FDR = Hotelling’s T squared test with Benjamini-Hochberg correction) (right). (C) ATAC-seq signal at PAR bZip footprints (DBP, HLF, and TEF) detected in both CON and CD-tRF samples and subsequently visualized in CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) samples at CT4 and CT16. Signal is normalized to reads per genomic content and is plotted ±0.5 kb from PAR bZip motif center. Shaded dark regions represent areas of accessible chromatin (top). Enriched GO: Biological Process pathways annotated from common PAR bZip footprints (DBP, HLF, and TEF) detected in both CON and CD-tRF. Key pathways associated with β cell secretory function are highlighted (bottom). TFBS, Transcription factor binding sites.