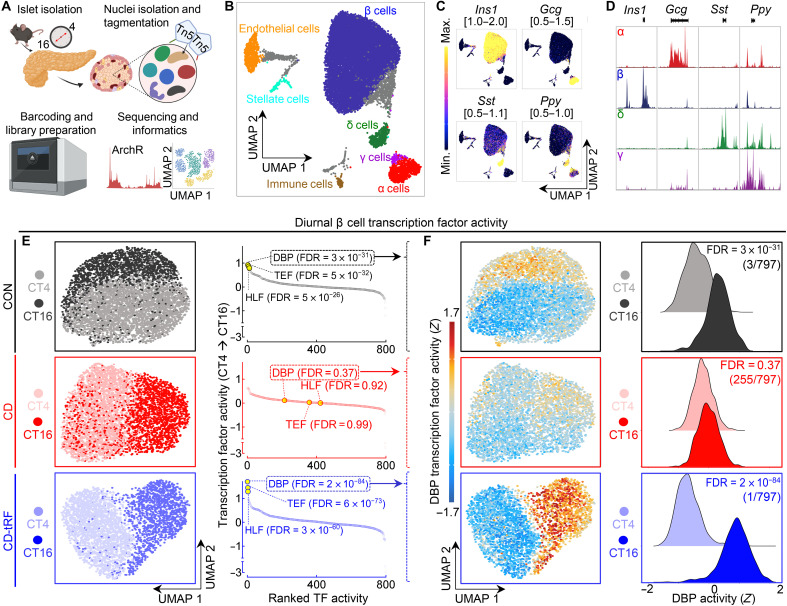
Fig. 6. tRF promotes diurnal PAR bZip transcription factor activity in single β cells under conditions of circadian disruption.
(A) Overview of study design. Islet cell nuclei were isolated from CON, CD, and CD-tRF mice at CT4 and CT16 and subjected to single-cell ATAC-sequencing (scATAC-seq). (B) Uniform Manifold Approximation and Projection (UMAP) of 16,470 islet cells with each dot representing one cell. Identified islet cell types are labeled (n = 2 mice per condition), and gray shaded cells are unidentified because of activity of multiple canonical islet cell markers. (C) UMAP of gene activity scores for canonical endocrine islet cell markers Insulin 1 (Ins1), Glucagon (Gcg), Somatostatin (Sst), and Pancreatic Polypeptide (Ppy). Gene activity score ranges are noted on top of each UMAP. Yellow represents high activity (open chromatin) at noted gene loci, while blue/purple represents low activity (closed chromatin). (D) Corresponding track plotting of pseudo-bulk chromatin accessibility at Ins1, Gcg, Sst, and Ppy gene loci in identified endocrine cells (β, α, δ, and γ cells). Gene loci are depicted below each genome-wide browser track. Signal is normalized to maximal accessibility of each loci across the displayed cell types. (E) UMAP of diurnal β cells under CON (gray/black), CD (pink/red), and CD-tRF (light blue/dark blue) conditions at CT4 and CT16 (left). Global diurnal changes in Z-normalized TF activity (out of 797 cis-BP TFs tested) in CON, CD, and CD-tRF β cells (FDR = Wilcoxon rank sum test with Benjamini-Hochberg correction) (right). Note that PAR bZip (DBP, HLF, and TEF) TFs are highlighted in yellow with corresponding FDR values presented in parentheses. (F) UMAP of Z-normalized diurnal β cell DBP activity in CON, CD, and CD-tRF β cells assessed using ChromVar at CT4 and CT16 (left). Histograms depicting diurnal differences in Z-normalized DBP activity in CON, CD, and CD-tRF β cells at CT4 and CT16 (right).