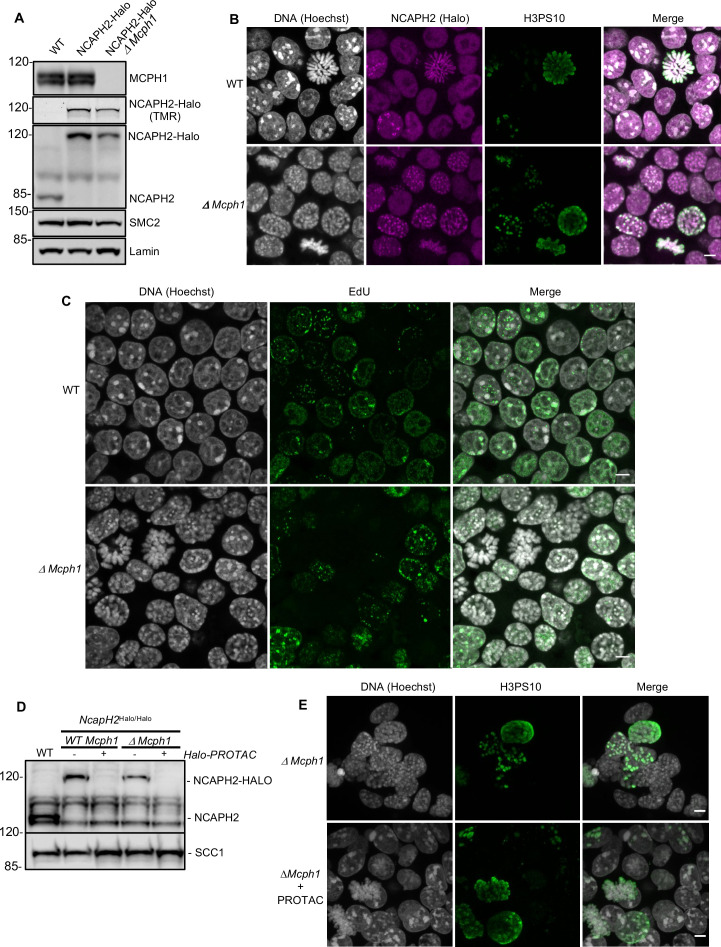
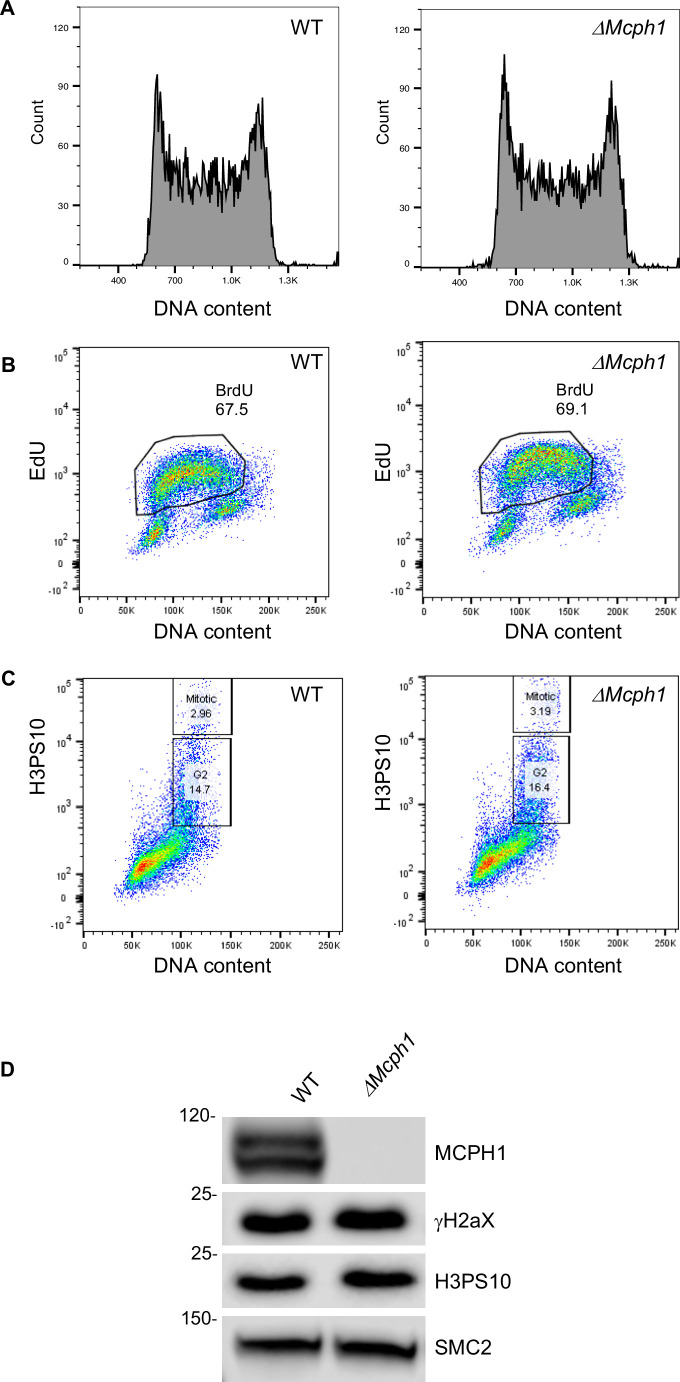
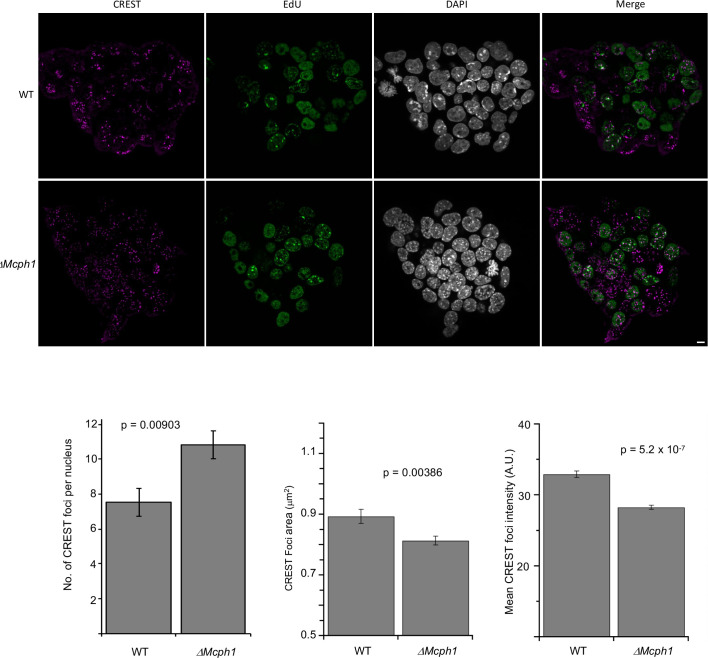
Figure 1. The deletion of Mcph1 in E14 cells induces condensin II-dependent chromosome condensation in both G1 and G2 phases of the cell cycle.
(A) In gel TMR-Halo detection and western blot analysis of E14 cells wild type, Ncaph2Halo/Halo and Ncaph2Halo/Halo Mcph1Δ/Δ. TMR signal detects NCAPH2-Halo tagged. The anti-NCAPH2 antibody shows that all the NCAPH2 protein expressed is fused to the Halo-tag and that the expression levels are similar to wild type but reduced after Mcph1 deletion. The anti-SMC2 detection shows similar levels of condensin in the three cell lines. (B) Immunofluorescence analysis of Histone H3 phosphorylated on serine 10 (green) combined with TMR detection of NCAPH2-Halo (Red) in Mcph1wt/wt and Mcph1Δ/Δ cells. The DNA organisation was analysed using Hoechst. (C) EdU incorporation in Mcph1wt/wt or Mcph1Δ/Δ cells.(D) Western blot analysis of Halo-PROTAC induced NCAPH2-Halo degradation in wild-type, Ncaph2Halo/Halo Mcph1wt/wt and Ncaph2Halo/Halo Mcph1Δ/Δ cells using an anti-NCAPH2 antibody. Anti-SCC1 was used as a loading control. (E) Immunofluorescence analysis of the chromosome decompaction induced by 16 hr treatment of Mcph1Δ/Δ cells with Halo-PROTAC. Immunofluorescence analysis of Histone H3 phosphorylated on serine 10 (green) was used to compare similar cell cycle stages. All the cells showed a diffuse chromatin organisation (150 cells counted). Scale bar, 5 μm.