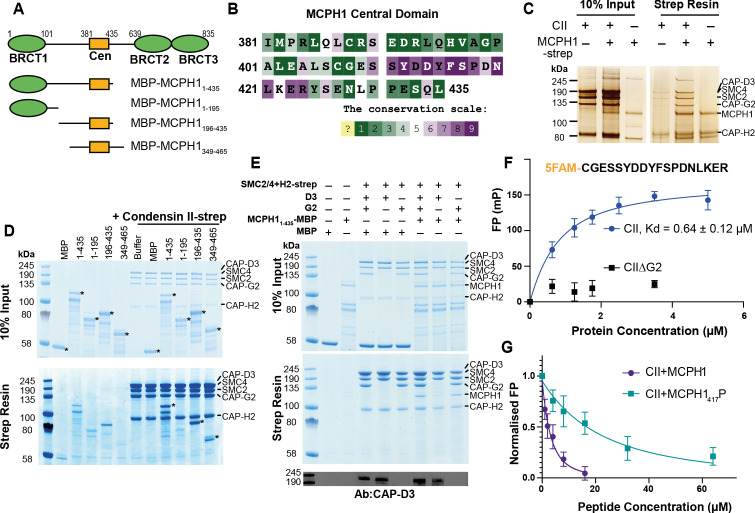
Figure 5. Human condensin II interaction with MCPH1.
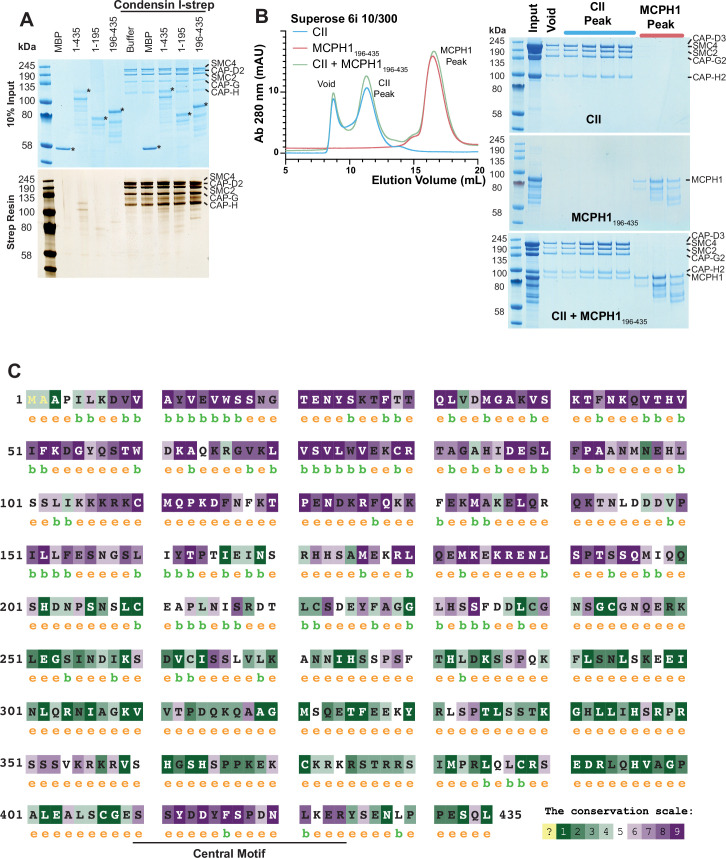
(A) Domain structure of MCPH1 and MBP fusion constructs that were expressed in E. coli and used in binding assays. BRCT domains are indicated in green and the central domain in yellow. (B) Conservation analysis of the MCPH1 central domain with the ConSurf server. (C) Strep tag pull-down indicating full-length MCPH1 binds condensin II. Full-length MCPH1 and condensin II were expressed in insect cells and separately purified, before being mixed on strep-tactin sepharose. Samples of input and resin after run on SDS page and visualised with silver stain. (D) Strep-tag pull-down assay indicating strep tagged condensin II pulls down MBP-MCPH1 constructs that contain the central domain, but not MBP-MCPH11-195 or MBP alone. SDS page gel visualised with Coomassie stain, (*) indicates the running position of the MBP/MCPH1 construct used. (E) Strep pull-down assay showing strep-tagged pentameric condensin II or tetrameric condensin II lacking NCAPD3 can pull down MBP-MCPH11-435, while tetrameric condensin lacking NCAPG2 does not pull down MCPH1. The lower panel shows a western blot performed using strep-resin samples, blotted using an anti-NCAPD3 antibody. (F) Fluorescence polarisation binding assay using 5-FAM-labelled MCPH1407-424 peptide and increasing concentration of either pentameric condensin or tetrameric condensin II lacking MCPH1 binding subunit NCAPG2 (CIIΔG2). (G) Peptide competition assay using a fixed concentration of 5-FAM labelled MCPH1407-424 and condensin II with an increasing amount of MCPH1407-424 wild-type or phosphorylated at serine 417. All error bars indicate standard deviation from three replicates.