Abstract
The direct oncolytic effect of Newcastle disease virus (NDV) depends on the following two aspects: the susceptibility of cancer cells to virus infection and the ability of virus itself to lyse cancer cells. First, we investigate the susceptibility of cancer cells to NDV infection, HepG2, MDA-MB-231, and SH-SY5Y cells were susceptible, A549, MCF7, and LoVo cells were less susceptible. To investigate the molecular mechanism responsible for cancer cell susceptibility, transcriptome sequencing was carried out. We found that the levels of alpha-sialic acid acyltransferase were upregulated in MDA-MB-231 cells compared with MCF7 cells, and the interferon was downregulated. Second, to optimize the oncolytic capacity of the wild-type rClone30, a series of chimeric viruses rClone30-Anh(HN), rClone30-Anh(F), and rClone30-Anh(HN-F) were constructed by exchanging the HN gene, F gene or both of non-lytic rClone30 strain with lytic strain Anhinga. rClone30-Anh(F) and rClone30-Anh(HN-F) enhanced the oncolytic effect of the rClone30, and this enhancement is more obvious in the susceptible cells. The oncolytic mechanism of rClone30-Anh(F) was analyzed by transcriptome analyses, in comparison with rClone30, rClone30-Anh(F) upregulated the expression of ATG5, Beclin 1, and MAP1LC3B, thus activating autophagy and promoting the production of syncytia. In conclusion, our study provides a strategy to enhance the oncolytic effect of rClone30.
Subject terms: Genetic vectors, Breast cancer
Introduction
Newcastle disease virus (NDV) is an avian paramyxovirus with a negative single-strand RNA genome and it has been shown to be a potent oncolytic agent with an attractive safety profile in humans in phase I and II clinical trials [1, 2]. NDV exhibits several advantages when compared with other oncolytic agents in development [3]. First, NDV is an avian pathogen, more than half century application of the virus as an avian vaccine demonstrates a super genome stability [4]. Second, serological studies indicated that ~96% of the human population is seronegative for NDV, avoiding the problem of preexisting immunity in humans, which is potential problems with vaccinia virus, herpes simplex virus, and adenovirus [5]. Last, a wide variety of cancers can be treated by NDV due to ubiquitous nature of the NDV receptor [6]. However, despite the advantages discussed above, results from clinical trials are not satisfactory [7–9].
The oncolytic effect of NDV attributes to two aspects, one is the oncolytic ability of virus per se and other is the susceptibility of host cancer cells to virus infection. Previous studies showed that NDV is specific to all cancer cell lines from ecto-, endo-, and meso-dermal origin [10] including the cells of colorectal, gastric, pancreatic, bladder, breast, ovarian, renal, lung, larynx, and cervical carcinomas, glioblastoma, melanoma, pheochromocytoma, lymphomas of different origins, fibrosarcoma, osteosarcoma, and neuroblastoma, but not normal cells [11–13]. However, our unpublished data showed the susceptibility of cancer cell lines to NDV infection was substantially different. We speculated that the susceptibility of different cancer cells to NDV infection may vary, it is necessary to conduct a systematic study on the susceptibility of cancer cells to NDV infection.
Another aim of the research is to enhance the oncolytic effect of existing NDV vectors via virus-engineering strategies. Numerous pieces of evidence indicate that the oncolytic capability of NDV in vivo is correlated to its ability to form syncytia in vitro [14]. The mesogenic strain Anhinga defined as lytic strain could be strong oncolytic virus due to induction of strong syncytia formation in vitro, while the lentogenic strain clone30 defined as non-lytic strain induces weak syncytia formation in vitro [15]. Many studies demonstrate that the hemagglutinin-neuraminidase (HN) and fusion (F) protein of NDV virion mediates the fusion of infected cells with their neighboring uninfected cells, production of syncytia and thus play essential roles in NDV-induced oncolytic effect [16, 17]. The F protein is a class I membrane protein present as a trimer in the virion, the cleavage site of the F protein is responsible for the major determinant of virulence and the formation of syncytia [18, 19]. The HN of NDV is a class II integral membrane protein, found as a tetramer in the virion, mediating receptor recognition of sialic acid at the end of host cell surface proteins [20]. Previous study showed that mutations at positions Asp198 and Arg174 reduced syncytial formation, suggesting that the HN gene plays a pivotal role in NDV-induced syncytial formation [21]. The interaction of HN and F proteins from the same viral strain is necessary for the most efficient function of receptor recognition and fusogenicity because the HN head domain carries the receptor-binding activities and the HN stalk domain harbors the site that determines the F protein specificity in promoting cell–cell fusion [22, 23]. Furthermore, previous study suggested that F and HN cooperatively disturb mitochondrial fusion–fusion homeostasis to enhance mitochondrial biogenesis, synergistically inducing significant syncytia formation accompanied with complete autophagic flux in DF-1 and A549 cells [24, 25]. But, the previous studies mainly focuses on the contributions of the F and HN proteins to pathogenicity and in vitro fusion activity, the roles of the F or HN on the oncolytic effect of NDV in vivo have yet to be studied.
Material and methods
Cell lines and cell cultures
Human lung epithelial carcinoma cells (A549), human breast cancer cells (MCF7), human neuroblastoma cells (SH-SY5Y), human colon cancer cells (LoVo), and African green monkey kidney cells (Vero) were purchased from ATCC. Human breast cancer cells (MDA-MB-231) was kindly offered by GS (College of Life Science, Northeast Forestry University). HepG2 cells which are perpetual cell line consisting of human liver carcinoma cells were purchased from China Peking Union Medical College (ATCC No. HB-8065). Mouse 4T1 breast cancer cells which is normally used to build a transplant-tumor mice model were kindly offered by GS (College of Life Science, Northeast Forestry University). BHK-21 cells were kindly offered by Prof. Karl-Klaus Conzenlmann (Max-von Pettenkofer Institut, Muenchen), Propagation of rescued viruses was performed on BHK-21 cell monolayers. Primary chicken embryo fibroblasts (CEF) were prepared from 9- to 11-day-old SPF chicken embryos. HepG2, MCF7, BHK-21, and CEF cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) FBS, 1% (v/v) penicillin/streptomycin. MDA-MB-231, SH-SY5Y, A549, and 4T1 were maintained in RPMI 1640 supplemented with 10% (v/v) FBS, 1% (v/v) penicillin/streptomycin. LoVo cells were maintained in F-12K medium supplemented with 10% (v/v) FBS, 1% (v/v) penicillin/streptomycin. Vero cells were maintained in minimum essential medium medium supplemented with 10% (v/v) FBS, 1.0 mM sodium pyruvate, and 0.1 mM nonessential amino acids. All cell lines were authenticated using Short Tandem Repeat profiling, tested for mycoplasma contamination and grown at 37 °C humidified incubator with 5% CO2 atmosphere.
Viruses
Parental viruses Anhinga/U.S.(FL)/44083/93 (GenBank: AY562986.1) was kindly offered by Southeast Poultry Research Laboratory. The recombinant lentogenic NDV clone30 strain, the recombinant mesogenic Anhinga strain (referred to here as rClone30, rAnh), the recombinant lentogenic clone30 expressing red fluorescence protein (referred to here as rClone30-RFP), and the recombinant mesogenic Anhinga expressing red fluorescence protein (referred to here as rAnh-RFP) were engineered and rescued as previously described [26]. All recombinant Newcastle disease viruses (rNDVs) were propagated in embryonated chicken eggs.
To assess the role of F and HN gene in oncolytic effect, a reverse genetic system was developed using the lentogenic NDV Clone30 strain to provide backbone for gene exchange. Briefly, plasmids pClone30 was linearized by digesting with the restriction enzymes to release the F gene or HN gene of the pClone30 plasmids for substitution with that from the pAnh plasmid. The overlap PCR amplified cDNA spanning the complete coding regions of the F or HN genes plus the surrounding sequences of the pClone30 were digested with the same restriction enzymes and ligated into the pClone30 backbone, named pClone30-Anh(HN), pClone30-Anh(F), and pClone30-Anh(HN-F), respectively (Table 1a, b). The recombinant plasmid was subjected to sequence analysis to confirm the fidelity of the PCR insertion, as well as the intergenic transcription start and stop sequences and the gene order. Then the recombinant NDV virus was rescued as described by Bai et al. [27], the recombinant plasmids were cotransfected with helper plasmids pBL-NP, pBL-P, and pBL-L (2000, 1000, 500, 250 ng, respectively) into BHK-21 cells stably expressing T7 RNA polymerase by Lipofectamine 3000 reagent. The virus was rescued and amplified by inoculation of the supernatant from the transfected cells into the allantoic cavity of specific-pathogen free chicken embryos. Viruses in HA-positive allantoic fluid were stored at −80 °C. After the second passage in eggs, viral RNA was extracted and the region containing the engineered molecular tag was amplified by RT-PCR using the forward primers and the reverse primers (P4, P5, P4a, and P5a, Table 1a, b). Then RT-PCR products were purified and sequenced to confirm that the rescued virus was generated from the infectious clone. The rescued viruses were named as rClone30-Anh(HN), rClone30-Anh(F), and rClone30-Anh(HN-F), respectively. The recombinant viruses rClone30, rClone30-RFP, and rClone30-Anh(HN) wee maintained in a solution containing 0.001% trypsin while the other viruses is not
Table 1.
The primers for HN gene amplification of moderately virulent strain and F gene amplification of moderately virulent strain, and primers of real-time PCR.
| a. The primers for HN gene amplification of moderately virulent strain | |
|---|---|
| Primer name | Primer sequence (5′−3′) |
| Primer 1(P1) | GGCCTGAGAGGCCTTCAGAGAGTTAAGA |
| SfiI | |
| Primer 2(P2) | GACTACATGATCCATGATTGAGGACTGTTGTCGGT |
| Primer 3(P3) | CAACAGTCCTCAATCATGGATCATGTAGTCAGCAG |
| Primer 4(P4) | ATGGATCATGTAGTCAGCAG |
| Primer 5(P5) | TTAAACCCTGTCTTCCTTGA |
| Primer 6(P6) | TTATAATTGACTCAATTAAACCCTGTCTTCCTTGA |
| Primer 7(P7) | AGACAGGGTTTAATTGAGTCAATTATAAAGGAG |
| Primer 8(P8) | CGTACGAATGCTGCTGAACT |
| BsiwI | |
| b. The primers for F gene amplification of moderately virulent strain | |
|---|---|
| Primer name | Primer sequence (5′−3′) |
| Primer 1a(P1a) | CACGTG AAAGCGCCAGAGAAGATTCCCGGGA |
| PmlI | |
| Primer 2a(P2a) | TTTGGGGCCCATCTTGCACCTGGAGGGCGCCAACCGG |
| Primer 3a(P3a) | TCCAGGTGCAAGATGGGCCCCAAACCCCCCACCGGAA |
| Primer 4a(P4a) | ATGGGCCCCAAACCC |
| Primer 5a(P5a) | TCATGTTTTTGTGGTGGCTC |
| Primer 6a(P6a) | CCTCATCTGTGT TCATGTTTTTGTGGTGGCTCTCATC |
| Primer 7a(P7a) | ACAAAAACATGAACACAGATGAGGAACGAAGGTTTCC |
| Primer 8a(P8a) | GGCCTGAGAGGCCACGCGTCGCCGCGGGCCGGTT |
| SfiI | |
| c. Primers of Real-time PCR | |
|---|---|
| Gene name | Primer sequence (5′−3′) |
| IFN-α-PF | GCCTCGCCCTTTGCTTTACT |
| IFN-α-PR | CTGTGGGTCTCAGGGAGATCA |
| IFN-β-PF | GCTTGGATTCCTACAAAGAAGCA |
| IFN-β-PR | ATAGATGGTCAATGCGGCGTC |
| IRF7-PF | GCTGGACGTGACCATCATGTA |
| IRF7-PR | GGGCCGTATAGGAACGTGC |
| IFIT1-PF | GCGCTGGGTATGCGATCTC |
| IFIT1-PR | CAGCCTGCCTTAGGGGAAG |
| Caspase-3-PF | CATGGAAGCGAATCAATGGACT |
| Caspase-3-PR | CTGTACCAGACCGAGATGTCA |
| Bax-PF | CCCGAGAGGTCTTTTTCCGAG |
| Bax-PR | CCAGCCCATGATGGTTCTGAT |
| Bcl2-PF | GGTGGGGTCATGTGTGTGG |
| Bcl2-PR | CGGTTCAGGTACTCAGTCATCC |
| ATG5-PF | AAAGATGTGCTTCGAGATGTGT |
| ATG5-PR | CACTTTGTCAGTTACCAACGTCA |
| Beclin 1-PF | GGTGTCTCTCGCAGATTCATC |
| Beclin 1-PR | TCAGTCTTCGGCTGAGGTTCT |
| LC3B-PF | AAGGCGCTTACAGCTCAATG |
| LC3B-PR | CTGGGAGGCATAGACCATGT |
Virus titration, ICPI, MDT assays, and determination of virus growth
Virus titration was performed by HA or HI test and the 50% tissue culture infective dose (TCID50) test on CEF cell monolayers. Viral titer in cell culture was calculated by the Reed and Muench [28]. To characterize the chimeric virus and parental virus’s pathotypes, two of the standard pathogenicity assays were performed: the mean death time (MDT) in embryonated chicken eggs and the intracerebral pathogenicity index (ICPI). The EID50 (50% egg infective dose) and MDT were performed by inoculating serial tenfold dilutions of the chimeras or parental virus stocks into 9 day embryonated chicken eggs. The ICPI test was performed by inoculation of infective allantoic fluid directly into the brain of 1 day SPF chickens. Calculation of the ICPI, EID50, and MDT were performed as previous study [29].
Viral growth was determined in the primary CEF. CEF cells were plated in six-well dishes at density of 3.0 × 105 cells per well and cells in six-well plates were infected with 0.1 MOI recombinant viruses. After incubation, cells were washed three times with PBS, and fresh medium was added. Cells with supernatants were frozen at the time indicated i.e., 12, 24, 48, and 72 post-infection. Intracellular virus titers were measured with a 50% tissue culture infective dose (TCID50) assay. Finally, a growth curve was drawn following the virus titer in different time.
Immunofluorescence assay and flow cytometry
HepG2, MDA-MB-231, SH-SY5Y, A549, MCF7, LoVo, and Vero cells were plated at 3.0 × 105 cells per well and infected with rClone30-RFP or rAnh-RFP at a MOI of 0.1. Fluorescence images were photographed using an inverted fluorescence microscope under ×100 magnification (Nikon, Japan)at 24 h pi, and the expression levels of red fluorescent protein (RFP) were analyzed by flow cytometry (BD, USA). Vero cells were used as a reference and the cells incubated with normal allantoic fluid were used as control.
RNA sequencing analysis
To assess the gene expression in the cells infected with NDV, RNA sequencing (RNAseq) was performed. After MDA-MB-231 and MCF7 cells were infected with allantoic fluid or recombinant viruses at a MOI of 0.1. RNA samples isolated from frozen cells using a Total RNA Extractor (Trizol) (#B511311, Sangon Biotech, Shanghai) were converted into cDNA libraries using the Illumina Hieff NGS™MaxUp Dual-mode mRNA Library Prep Kit (Illumina #12301ES96, YEASEN), and then RNAseq was performed. Differentially expressed genes were analyzed with a DEGseq algorithm by Sangon Biotech.
Real-time PCR
Total RNA extraction and reverse transcription to cDNA (1 µg RNA) were performed as previously described [30] and quantitative real-time PCR (qRT-PCR) performed with 50 ng cDNA, 100 nmol/L of each primer and the IQ-SYBR-green Mastermix (Bio-Rad, Milano, Italy) was used to quantify mRNA levels of a gene using the ABI-Prism 7300 (Applied Biosystems; ABI). Gene-specific primers were used in each qRT-PCR reaction (Table 1c) and all qRT-PCR assays were performed in triplicates in a 96-well plate according to the manufacturer’s protocol. The results were normalized against β-actin and expressed as fold changes in relative mRNA expression level using the 2−ΔΔCT method [31].
Cell viability assay
A short-term microculture tetrazolium (MTT) assay was used to quantify cell viability. A total of 1 × 104 cells were plated per well in a 96-well plate and incubated with virus at 0.01, 0.1, 1, and 10 MOI. Twenty microliters MTT solutions (5 mg/ml in sterile phosphate-buffered saline) were added to the cell 48 h after infection. The MTT solution in the wells was discarded after 4 h incubation, and 150 μl DMSO was added, then the plates were vibrated for 10 min, a spectrophotometer was used to determine the optical density (OD) at the absorbance of 570 nm, all samples were analyzed in triplicates. Cytotoxicity was quantified as the difference in cell viability between the experimental samples and the uninfected controls. The cell viability was converted and expressed using the formula:
Inhibition ratio = (control group OD − treatment group OD)/control group OD × 100%.
Fusion promotion
To assess the difference of the fusion capability of chimeric virus versus rClone30 in different cancer cell lines, HepG2, MDA-MB-231, MCF7, A549 H22, and 4T1 cells monolayers in six-well plate were infected with recombinant virus at the 0.1 MOI, respectively. Cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) for localization of nuclei after 24 h post-infection, and the representative images were captured under ×100 magnification with an camera (Nikon, Japan) attached to a microscope.
Annexin V-FITC and PI binding assay
Apoptosis was determined by translocation of phosphatidylserine to the cell surface using an Annexin V-FITC and PI apoptosis detection kit (Nanjing KeyGen Biotech. Co. Ltd, China). Overall, 0.1 MOI recombinant viruses were added into the six-well plate lined with HepG2, MCF7 cells, the cells were harvested and washed twice with cold PBS after 24 h of incubation, and resuspended in Annexin V-FITC and PI keeping in dark for 30 min. Cells were analyzed in a flow cytometry using FL1 (530 nm) band pass filters of FACS Calibur (BD, USA), and data were analyzed by the Cell Quest software (BD, USA).
Establishment of subcutaneous tumor-bearing model
All procedures involving animals followed the guidelines issued by National Institute of Health and the Institutional Animal Care and Use Committee of Northeast Agriculture University. Healthy 6-week-old female BALB/c mice with a weight of 19 ± 1 g were permitted to feed and drink freely. H22 cells or 4T1 cells (1 × 106) were subcutaneously implanted into the right groin of mice and tumors were allowed to grow until the average diameter reached 5–8 mm. Mice were randomly divided into different groups (n = 16), and intratumorally injected with allantoic fluid or 1 × 108 pfu of the indicated viruses every day. Tumor volumes were calculated using the following formula: tumor volume (V) = 4/3 × π × S2/2 × L/2, where S is the smallest measured diameter and L is the largest diameter. Animals were humanely culled when tumor size reached 18 mm in any dimension or at defined experimental time points. After the treatment, six animals of each group were sacrificed, their tumors and spleens were excised, weighted, frozen, or immersed in 4% paraformaldehyde. The remained mice in each group were continually monitored till 100 days to observe the survival rate.
Histological analysis and TUNEL assay
Hematoxylin and eosin (H&E) staining was carried out following the standard protocol. Briefly, tissue specimens were first fixed in 4% PBS-buffered formaldehyde, and then embedded in paraffin. Tissue sections of 4–5 μm were finally stained with H&E solutions and prepared for the TUNEL assay (terminal deosynucleotidy transferase-mediated dUTP nick and labeling assay). The Apoptosis of tumor cell was detected by the TUNEL apoptosis assay kit (Beyotime, #C1098).
Isolation of lymphoid cells and detection of Immune cell populations
After all the mice were killed, their spleens were immediately placed into a plate that contained PBS with 2% heat-inactivated FBS and dissected mechanically. Single-cell suspensions were prepared by gently teasing them through sterile stainless-steel screens to eliminate clumps and debris [32]. The erythrocytes were lysed in red blood cell lysis buffer that consisted of 0.155 M NHCl, 0.01 M KHCO3, and 0.1 mM EDTA. The remaining cells were washed three times with a large volume of RPMI 1640. All preparation procedures were performed at 4 °C. After the last centrifugation, the cells were counted using a hemocytometer and resuspended in an appropriate volume of PBS. Single-cell suspensions of spleen from the mice were incubated with PE anti-mouse CD4+ (Clone: REA604, miltenyibiotec), APC anti-mouse CD8+ (Clone: REA601, miltenyibiotec) FITC anti-mouse CD3+ (Clone: REA641, miltenyibiotec) at room temperature for 2 h in the dark. After incubation, the cells were washed twice with PBS and analyzed by a flow cytometer (BD Bioscience, San Jose, CA).
Animal safety assessment
Recombinant viruses rClone30-RFP or rAnh-RFP was intravenously injected into subcutaneous tumor-bearing mice or normal mice to observe the distribution of virus. All the viruses mentioned above (rClone30, rClone30-Anh(HN), rClone30-Anh(F), and rClone30-Anh(HN-F)) were intraperitoneally injected into normal mice for 30 days, serum was prepared from whole blood for determination of serum concentrations of AST, ALT, BUN, and creatinine.
Statistical analysis
The statistical significance of quantitative data between different groups was determined with GraphPad prism software (Version 5.01, GraphPad Software Inl., La Jolla, California). All data were expressed as mean ± SEM (standard error of the mean), significance was determined by performing one- or two-sided Student’s t tests and defined as a p value < 0.05, p value < 0.01.
Results
Generation of recombinant viruses
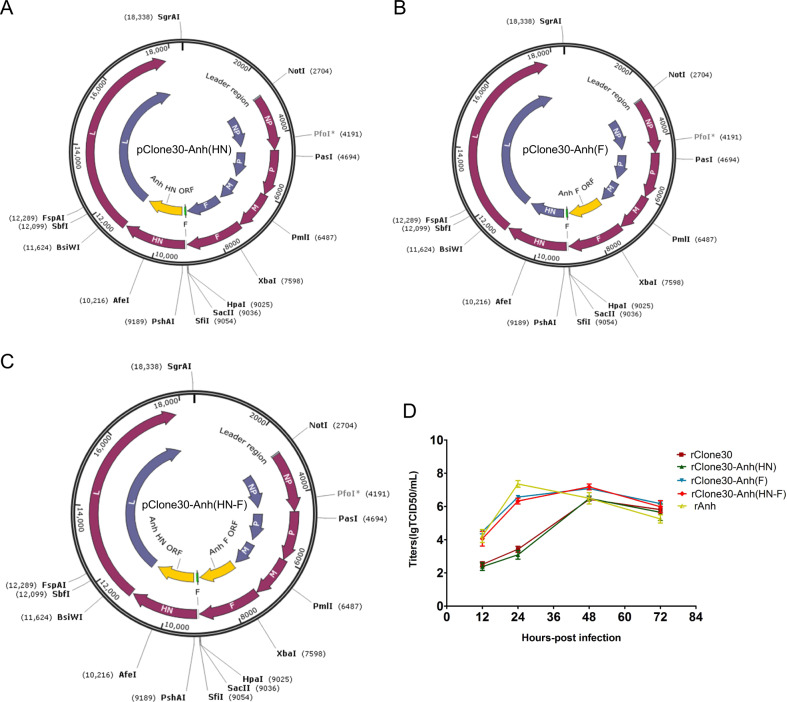
Chimeric viruses with the complete anti-genome sequence of the NDV were generated as described in the “Material and methods” section, and the HN, F gene of rClone30 has been exchanged with that of the Anhinga strain (Fig. 1a–c). The reverse genetics technology was used to generate viable infectious virus in BHK-21 cells and the HN and F genes in rescued viruses were verified by sequencing as described in the “Material and method” section.
Fig. 1. Schematic representation of the genomes and characterization of recombinant viruses.
The F gene, HN gene fragment or both of pClone30 were exchanged with that of Anhinga by overlap PCR (yellow), including the chimeric virus expressing the HN gene (a rClone30-Anh(HN)), F gene (b rClone30-Anh(F)) or both (c rClone30-Anh(HN-F)) of Anhinga virus. d Growth curves of recombinant Newcastle viruses. Primary chicken embryo fibroblasts cells (CEF) were infected with rClone30, rClone30-Anh(HN), rClone30-Anh(F), rClone30-Anh(HN-F), and rAnh at MOI of 0.1. Cell monolayers were lysed at 12, 24, 48, and 72 h post-infection for intracellular titer measurement by TCID50 analysis.
Test of virus titer and pathogenicity of the recombinant viruses
A summary of virus titer and the pathogenicity assay is shown in Table 2. HA test results showed that the HA titer of rClone30-Anh(F) was similar to parental strain (29) and the HA titer of rClone30-Anh(HN) and rClone30-Anh(HN-F) was lower than rClone30(27). The MDT observed for the rAnh was 82 h, while the rClone30-Anh(F) and rClone30-Anh(HN-F) was 96 and 92 h, respectively. The ICPI results for the rAnh, rClone30-Anh(F), and rClone30-Anh(HN-F) were 1.47, 1.17, and 1.21, respectively. The pathogenicity of rClone30-Anh(F) and rClone30-Anh(HN-F) were lower than that of the parental Anhinga strain (Table 2); the data showed that the TCID50 of rClone30-Anh(F) and rClone30-Anh(HN-F) were in coincidence with the Anhinga (Table 2). As shown in Fig. 1d, the chimeric viruses rClone30-Anh(F) and rClone30-Anh(HN-F) showed a higher titer than rClone30 in 12 and 24 h, and all viruses except rAnh have almost reached their maximum titer in 48 h. The TCID50 of rClone30 and rClone30-Anh(HN) was significantly lower than that of rClone30-Anh(F) and rClone30-(HN-F) at 12 and 24 h may be explained by assuming that the limited cell–cell spread of viruses that lack the Anhinga F gene compared with viruses that do contain this gene, due to differences in cleavage of the F proteins. But, there was no significant difference between rClone30 and chimeric virus rClone30-Anh(HN) in growth. These results indicate that the replacement of HN gene dose not influence the growth kinetics of rClone30 and the replacement of F gene enhance the fusogenic property of rClone30.
Table 2.
Comparison of virus titer and pathogenicity.
| Virus | HA | TCID50 | EID50 | MDT | ICPI |
|---|---|---|---|---|---|
| rClone30 | 29 | 2.09 × 107 | 6.67 × 109 | >120 h | 0.05 |
| rClone30-Anh(HN) | 27 | 1 × 107 | 1 × 109 | >120 h | 0.08 |
| rClone30-Anh(F) | 29 | 1.62 × 108 | 3.16 × 109 | 96 h | 1.17 |
| rClone30-Anh(HN-F) | 27 | 1.1 × 108 | 1 × 109 | 92 h | 1.21 |
| rAnh | 27 | 3.24 × 107 | 4.27 × 108 | 82 h | 1.47 |
The susceptibility of the selected cancer cells to NDV infection was different
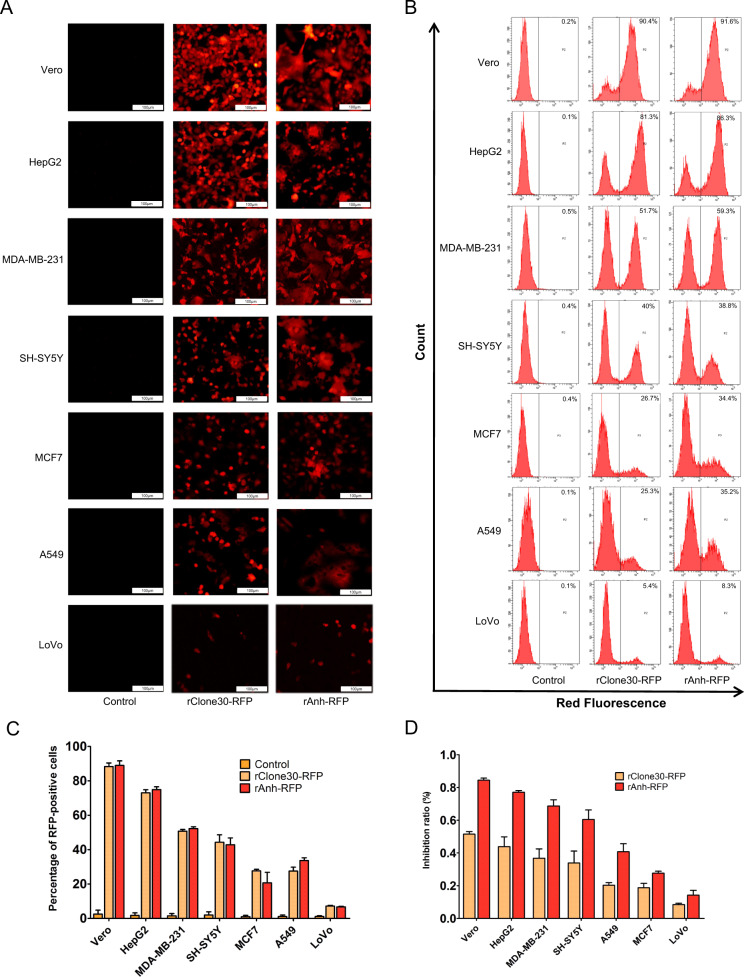
To investigate the susceptibility of cancer cells to NDV infection, Vero cells which is susceptible to NDV infection was used as a reference. The expression levels of RFP were detected by fluorescence microscopy and flow cytometry after the HepG2, MDA-MB-231, SH-SY5Y, A549, MCF7, and LoVo cells were infected with rClone30-RFP or rAnh-RFP at 0.1 MOI, respectively. At 24 h post-infection, red fluorescence was observed in these cells except for the mock-infected cells (Fig. 2a). The fluorescence intensity generated by rClone30-RFPs or rAnh-RFP was higher in HepG2, SH-SY5Y, and MDA-MB-231 cells than that in A549, MCF7, and LoVo cells. The HepG2 cells infected with rClone30-RFPs or rAnh-RFP displayed the highest red fluorescence intensity compared with other cell lines while the LoVo cells showed the lowest. The red fluorescence intensity of HepG2 cells was tenfolds than that of LoVo cells. To detect whether the susceptibility of cancer cells was different due to the different NDV strains, the red fluorescence intensity of the selected cancer cells infected with rClone30-RFP or rAnh-RFP was observed. The result showed that there was no significant difference between rClone30-RFP and rAnh-RFP (P > 0.05, Fig. 2b, c). Furthermore, the direct cytotoxic activity of rClone30-RFP and rAnh-RFP were tested in HepG2, MDA-MB-231, SH-SY5Y, A549, MCF7, and LoVo cells by a short-term MTT assay. As shown in Fig. 2d, rClone30-RFP and rAnh-RFP induced cell death at 48 h post-infection, HepG2, SH-SY5Y, and MDA-MB-231 cells was dramatically inhibited compared with A549, MCF7, and LoVo cells, the chimeric virus rAnh-RFP was superior to rClone30-RFP in suppressing the growth (p < 0.05). The susceptibility of cancer cells to NDV infection was categorized into three groups by analyzing the red fluorescence intensity and the growth inhibition rate: HepG2, SH-SY5Y, and MDA-MB-231 cells were more susceptible, A549 and MCF7 were moderately susceptible, LoVo cells were less susceptible. These results suggest that the susceptibility of cancer cells to NDV infection is different, and cells from the same organ also demonstrated different susceptibility like the breast cancer cells MDA-MB-231, which was more susceptible to NDV infection than MCF7 cells.
Fig. 2. The susceptibility of cancer cells to NDV infection.
a–c rClone30-RFP or rAnh-RFP at MOI of 0.1 were used to infect Vero cells and cancer cells in six-well plate. the fluorescence in the infected cells were detected and digitally photographed by fluorescence microscopy at ×100 magnification at 24 h (a), and analyzed using flow cytometry (b), the results of RFP fluorescence intensity were expressed as the percentage of RFP positive cells with SEM (*P < 0.05; **P < 0.01) (c). d Cytotoxicity effects of recombinant viruses on Vero cells and six cancer cells, the cells incubated with normal allantoic fluid were used as control. MTT method was used to detect the cell density at 490 nm. The data are the means ± SEM of triple samples (*P < 0.05; **P < 0.01), Vero cells were used as a reference.
The mechanism responsible for cancer cell susceptibility to NDV infection
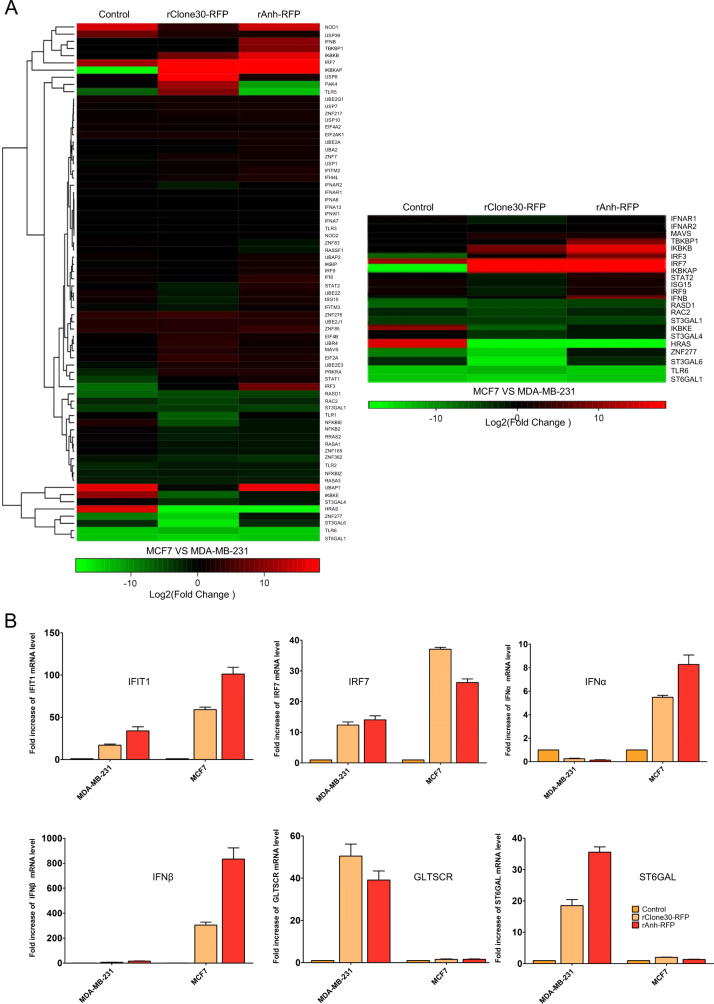
To explore the mechanism responsible for cancer cell susceptibility to NDV infection, transcriptome sequencing was used to analyze differentially expressed genes in susceptible MDA-MB-231 and less susceptible MCF7 cells infected with rClone30-RFP or rAnh-RFP. The results showed that many genes were differentially expressed after MDA-MB-231 and MCF7 cells infected with the viruses. We found that alpha-sialic acid acyltransferase, Rac2, and HRAS were downregulated and the expression of interferon (IFN) was upregulated in less susceptible MCF7 cells compared with that in susceptible MDA-MB-231 cells (greater than twofold change). Furthermore, real-time PCR assay confirmed this result, the levels of IFN-α, IFN-β, IRF7, and IFIT1 in MCF7 cell infected with rClone30-RFP or rAnh-RFP were significantly upregulated, while the level of Glioma tumor suppressor candidate region gene 2 (GLTSCR2) attenuated IFN-β and ST6GALI associated with the formation of α2,6-linked sialic acid was significantly downregulated (Fig. 3). These results suggest that the molecular mechanism which is responsible for cancer cell susceptibility to NDV infection was associated with the expression of IFN and α2,6-sialyltransferase.
Fig. 3. Alpha-sialic acid were upregulated and interferon were downregulated in susceptible cells.
a Heat map of differentially expressed genes from MDA-MB-231 or MCF7 infected with rClone30-RFP and rAnh-RFP, respectively. b Total RNA was extracted and analyzed by RT-qPCR with primer sets against the following transcripts: IRF7, IFIT1, IFN-α, IFN-β, GLTSCR2, and ST6GALI. The mRNA transcription level is normalized to that of β-actin and is expressed as the fold change relative to growing cells. The cancer cells incubated with normal allantoic fluid were used as control. Values are means ± SEM of two independent samples with triplicate qPCR.
rClone30-Anh(F) and rClone30-Anh(HN-F) are more fusogenic in susceptible cells than in less susceptible ones
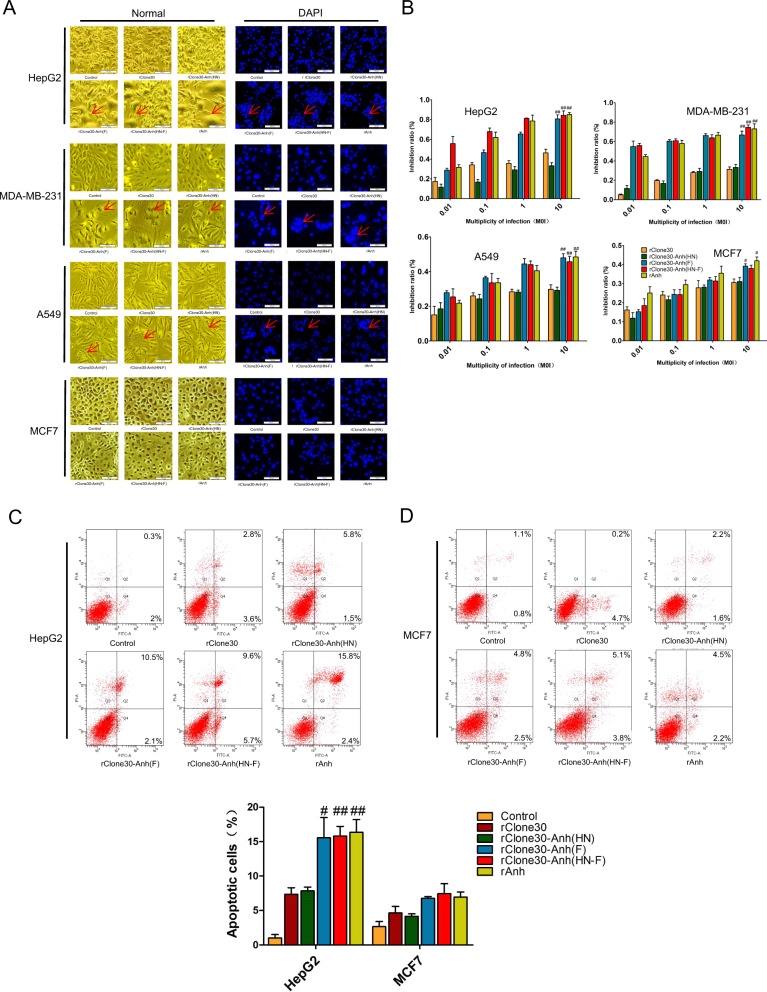
The susceptible cell lines HepG2, MDA-MB-231 and less susceptible cell lines A549, MCF7 were used to detect the fusogenic ability of chimeric virus in vitro. The results showed that more syncytia generated by rClone30-Anh(F) and rClone30-Anh(HN-F) compared with rClone30 and rClone30-Anh(HN) after infection with 0.1 MOI of viruses for 24 h in susceptible cell lines. However, in less susceptible cell lines, little syncytia was observed (Fig. 4a). The above results indicate that rClone30-Anh(F) and rClone30-Anh(HN-F) are more fusogenic in susceptible cells than in less susceptible ones, and rapid induction of cell–cell fusion, leading to extensive syncytial spread, is the mechanism of rClone30-Anh(F) and rClone30-Anh(HN-F)-mediated cell killing.
Fig. 4. Chimeric viruses expressing the F gene of lytic strain Anhinga induces serious cell death in vitro.
a Syncytia promotion analysis of different cancers. Cancer cells (HepG2, MDA-MB-231, A549, and MCF7) were infected with recombinant viruses at MOI of 0.1, and photomicrographs of cell monolayers were captured at 24 h post-infection (left). Cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) for localization of nuclei (right). Representative fields of view were shown at ×100 magnification. The cancer cells incubated with normal allantoic fluid were used as control. b Cytotoxicity effects of recombinant viruses on cancer cells. The cancer cells HepG2, MDA-MB-231, A549, and MCF7 cells incubated with normal allantoic fluid were used as control. MTT method was used to detect the cell density at 490 nm. The data are the means ± SEM. of triple samples (*P < 0.05; **P < 0.01). c, d NDV induce more apoptotic cells in susceptible cells. Apoptosis cells were quantified by flow cytometry with Annexin V-FITC and PI. Representative scatter plots of PI (y-axis) versus FITC (x-axis). Values represent mean ± SEM. with three replicates. *P < 0.05, **P < 0.01, compared with control. c HepG2. d MCF7.
rClone30-Anh(F) and rClone30-Anh(HN-F) are potent in suppression of cancer cell growth in vitro
The direct cytotoxic activity of the recombinant viruses was tested in HepG2, MDA-MB-231, A549, and MCF7 cells. As shown in Fig. 4b, all rNDVs induced cell death in a dose-dependent manner at 48 h post-infection, and the growth inhibition rate of rClone30-Anh(F) and rClone30-Anh(HN-F) was significantly higher than that of rClone30 and rClone30(HN) (p < 0.05). The growth inhibition was more obvious in the susceptible cells (HepG2, MDA-MB-231).
Apoptosis is enhanced by rClone30-Anh (F) and rClone30-Anh(HN-F) in both susceptible HepG2 cells and less susceptible MCF7 cells
To detect the sensitivity of tumor cells to virus-induced apoptosis, susceptible cells, and less susceptible cells (HepG2, MCF7) were infected with the recombinant viruses for 24 h, and the percentage of apoptotic cells was determined by the Annexin V-FITC and PI apoptosis detection kit. The Q2 and Q4 represent late apoptotic cells and early apoptotic cells, respectively. As shown in Fig. 4c, there was detected apoptosis in the two cancer cell lines (HepG2 and MCF7) and the apoptotic rate of the HepG2 cells markedly increased compared with MCF7 cells. After HepG2 cells were infected with rNDVs at 0.1 MOI for 18 h, the apoptotic percentage of rClone30, rClone30-Anh(HN), rClone30-Anh(F), and rClone30-Anh(HN-F) was 7.35 ± 0.95%, 7.85 ± 0.55%, 15.55 ± 2.5%, and 15.8 ± 1.5%, respectively. These results indicate that rClone30-Anh(F) and rClone30-Anh(HN-F) were more effective in inducing apoptosis in the HepG2 cells and there was no significant difference between the two viruses (Fig. 4c).
rClone30-Anh(F) induces cell death by the initiation of autophagy
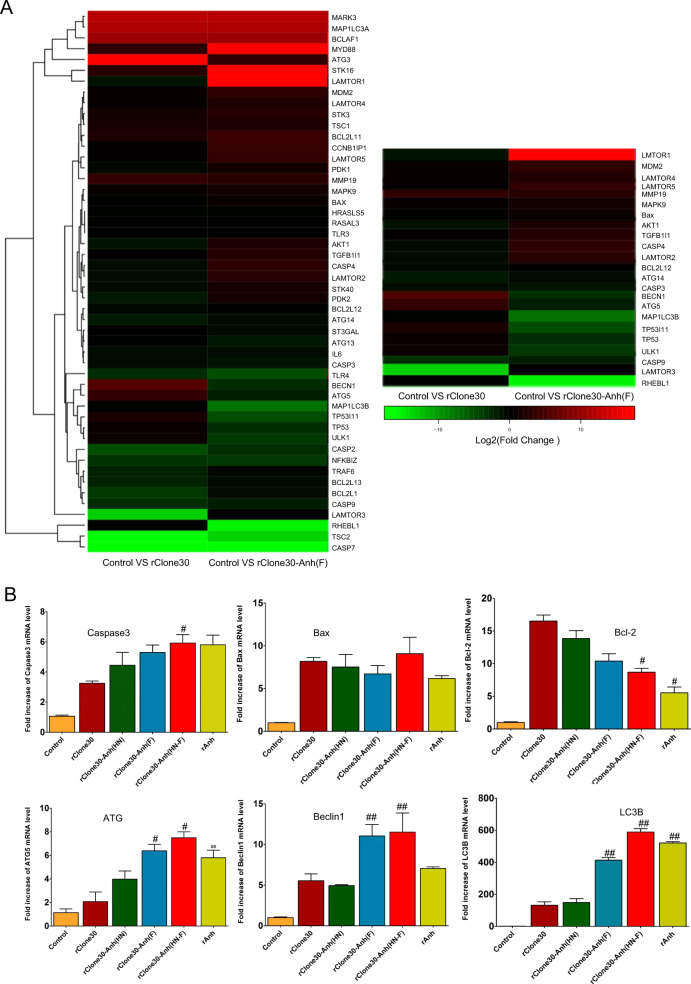
To elucidate the oncolytic mechanism of rClone30-Anh(F), the differential gene expression of MDA-MB-231 cells infected with rClone30 or rClone30-Anh(F) was analyzed by transcriptome sequencing. As shown in Fig. 5a, Using a fold change >2 as criteria, we found that the expression of many genes was changed after MDA-MB-231 cells was infected with rClone30 or rClone30-Anh(F). The result showed that 297 genes were similarly affected, but the other 232 genes were changed only in MDA-MB-231 infected with rClone30-Anh(F) (Supplementary Fig. 1). The apoptosis-related genes Caspase-3, Caspase-7, Caspase-9, BAX, and BCL2 were similarly affected in MDA-MB-231 cells with two viruses infected. However, Autophagy-related genes, AKT1, LAMTOR1, LAMTOR2, LAMTOR4, and LAMTOR5 were downregulated, TP53, ATG5, BECN1, ULK1, and MAP1LC3B were upregulated only in MDA-MB-231 cells infected with rClone30-Anh(F) (Fig. 5a). The expression of Caspase-3, BAX, BCL2, ATG5, BECN1, and MAP1LC3B was confirmed by RT-qPCR (Fig. 5b). These results suggest that rClone30-Anh(F) induced the syncytia formation is associated with autophagy.
Fig. 5. rClone30-Anh(F) upregulated the autophagy-related genes.
a The breast cancer cells MDA-MB-231 infected with rClone30 or rClone30-Anh(F) were harvested to carried out transcriptome analysis, and form a heat map of genes comprising apoptosis and autophagy. b The MDA-MB-231 cells infected with recombinant viruses were collected and total RNA were extracted. The mRNA transcription level of caspase-3, Bax, Bcl2, ATG5, Beclin 1, and LC3B were measured by Real-time PCR. Data represent mean ± SEM of triplicate samples (#P < 0.05, ##P < 0.01,compared with rClone30 group).
rClone30-Anh(F) and rClone30-Anh(HN-F) show a stronger oncolytic effect in H22-induced xenograft than 4T1-induced xenograft
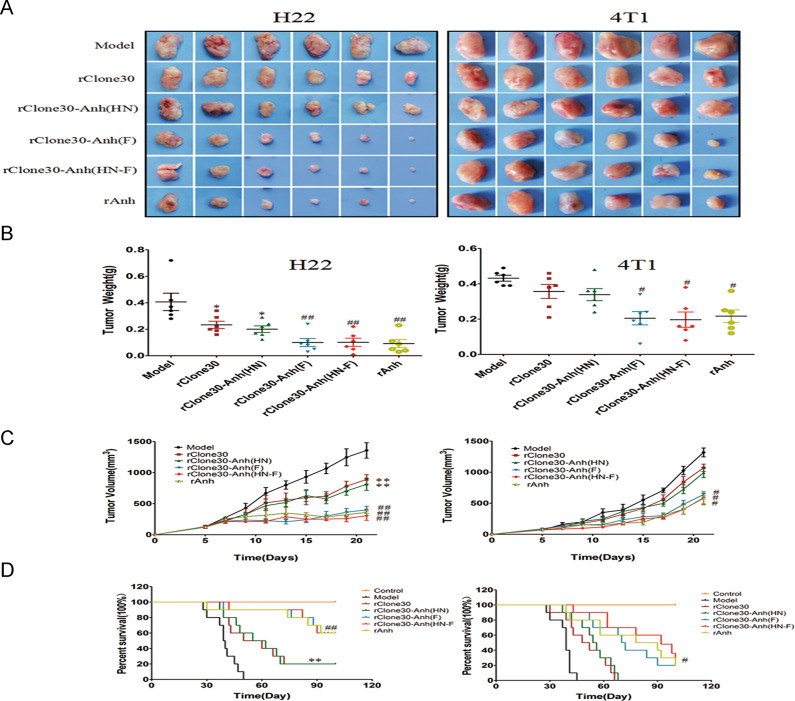
After mice were injected with susceptible H22 cells or less susceptible 4T1 cells, their volume and weight of tumors were recorded every 2 days, as shown in Fig. 6, compared with the model group, the average tumor volume and the average tumor weight decreased in all viral treatment groups. In the H22 tumor-bearing model, the average tumor size for the rClone30 group was 891.04 ± 75.73 mm3, the average tumor size for the rClone30-Anh(HN) group was 812.5 ± 98.02 mm3, while 400.91 ± 57.74 mm3, 307.5 ± 75.71 mm3, noted for the rClone30-Anh(F) and rClone30-Anh(HN-F) group (Fig. 6c). The average tumor weight for rClone30 and rClone30-Anh(HN) groups was 0.23 ± 0.027 g and 0.2 ± 0.033 g compared with 0.1 ± 0.03 g and 0.09 ± 0.03 g for rClone30-Anh(F) and rClone30-Anh(HN-F) groups, respectively (Fig. 6b). After necropsy, tumors excised from the model group were fairly large in size, the tumors in rClone30 and rClone30-Anh(HN) groups decreased slightly and tumors isolated from the rClone30-Anh(F) and rClone30-Anh(HN-F) groups were generally small (Fig. 6a). Furthermore, the survival rate of the tumor-bearing mice was also recorded. Over the 100 days (Fig. 6d), all ten mice in the model group were died, tumors of remaining 8/10 mice in the rClone30 and rClone30-Anh(HN) groups developed significant size and needed to be sacrificed. While only 4/10 mice in the rClone30-Anh(F) group and rClone30-Anh(HN-F) group developed tumors that required to be sacrificed. The overall survival of the mice in the long-term study was 0% for the model group, 20% for rClone30 and rClone30-Anh(HN) group, 60% for rClone30-Anh(F) group and rClone30-Anh(HN-F) group. 4T1-bearing mice treated with chimeric viruses rClone30-Anh(F) and rClone30-Anh(HN-F) showed a slightly suppression in tumor volume compared with the rClone30 group and rClone30-Anh(HN) group (P < 0.05) and there was no significant difference between rClone30-Anh(F) and rClone30-Anh(HN-F) groups. After experimental period, 10/10 mice in the model group, rClone30 group and rClone30-Anh(HN) group died, 8/10 mice in the rClone30-Anh(F) and rClone30-Anh(HN-F)groups died (Fig. 6). Remaining mice in rClone30-Anh(F) and rClone30-Anh(HN-F) groups also developed significant size, which the average diameter was 17.77 and 16.85 mm, respectively. These results suggest that the oncolytic effect of NDV on less susceptible cells 4T1-derived xenograft is inferior to susceptible cells H22-derived xenograft.
Fig. 6. Chimeric viruses expressing the F gene of lytic strain Anhinga show enhanced oncolytic effect in vivo.
Six-week-old female BALB/c mice were injected in the right groin with 106 H22 cells or 4T1 cells. Seven days post-infection the tumors were intratumorally treated with allantoic fluid, 1 × 108 pfu of recombinant Newcastle disease virus. a Tumor Tissues. b Tumor Weight. c The tumor volume of H22 (left) or 4T1 (right) tumor-bearing mice was measured every other day using digital calipers in two dimensions. d Survival of H22 (left) or 4T1 (right) models animal in 100 days period after treatment. The tumor-bearing mice were sacrificed when the tumor volume grew to a significant size (diameter > 18 mm). Control: untreated mice. Model: mice treated with allantoic fluid. *P < 0.05, **P < 0.01, compared with the Model group, #P < 0.05, ##P < 0.01, n = 6, compared with the rClone30 group.
rClone30-Anh(F) and rClone30-Anh(HN-F) enhance T-cell infiltration and apoptosis in the xenograft of tumor-bearing mice
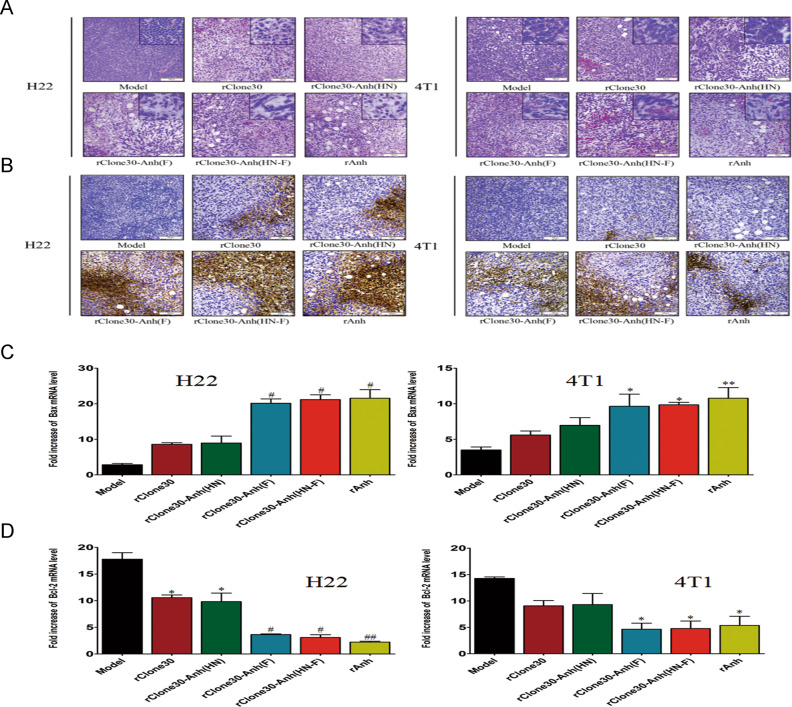
To illustrate the histopathologic changes of tumors infected with recombinant viruses, H&E staining was performed. The H&E staining pattern of the rClone30-Anh(F) and rClone30-Anh(HN-F)-infected xenografts exhibited more tumor-infiltrating lymphocytes and necrotic cells compared with rClone30 and rClone30-Anh(HN)-infected xenografts in both H22 and 4T1-bearing mice (Fig. 7a). Then we evaluated the cytotoxic effect of the recombinant virus on tumor tissue by TUNEL assay. Compared with the model group, we found an increase of the apoptotic cells in the rClone30 and rClone30-Anh(HN)-infected tumors in H22-bearing mice, but not in 4T1-bearing mice. rClone30-Anh(F) and rClone30-Anh(HN-F) infection resulted in more apoptotic cells in both H22 and 4T1-bearing mice, this effect was more obvious in the tumors of H22-bearing mice (Fig. 7b). Real-time PCR showed that the transcriptional levels of Bax significantly increased and the transcriptional level of Bcl2 decreased in the rClone30-Anh(F) and rClone30-Anh(HN-F) groups compared with rClone30 and rClone30-Anh(HN) groups in the H22 tumor tissues. However, in the 4T1 tumor tissues, the transcriptional levels of Bax / Bcl2 of rClone30-Anh(F) and rClone30-Anh(HN-F) groups increased a little (p < 0.05, Fig. 7c, d). These results suggest that rClone30-Anh(F) and rClone30-Anh(HN-F) is stronger than rClone30 and rClone30-Anh(HN) in T-cell infiltration and apoptosis of the xenografts and this effect is more obvious in H22-bearing mice.
Fig. 7. Chimeric viruses expressing the F gene of lytic strain Anhinga induce more T-cell infiltration, necrotic foci, and apoptotic cells in vivo.
a H&E staining of H22 tumor sections (left) and 4T1 tumor sections (right) after treatment of viruses. Representative fields of view were shown at ×100 magnification and ×400 magnification. b TUNEL assay (terminal deosynucleotidy transferase-mediated dUTP nick and labeling assay) was used to detect the apoptosis of tumor cells and the results were shown in brown. The larger the brown area, the more serious the apoptosis of cancer cells was. Representative fields of view were shown at ×100 magnification. c, d Transcriptional levels of Bax and Bcl2 genes in the tumor of H22 (left) and 4T1(right) tumor-bearing mice were calculated relative to the β-actin. Control: untreated mice. Model: mice treated with allantoic fluid. *P < 0.05, **P < 0.01, compared with the Model group. #P < 0.05, ##P < 0.01, n = 3, compared with the rClone30 group.
rClone30-Anh(F) and rClone30-Anh(HN-F) infection resulted in systematic T lymphocytes accumulation
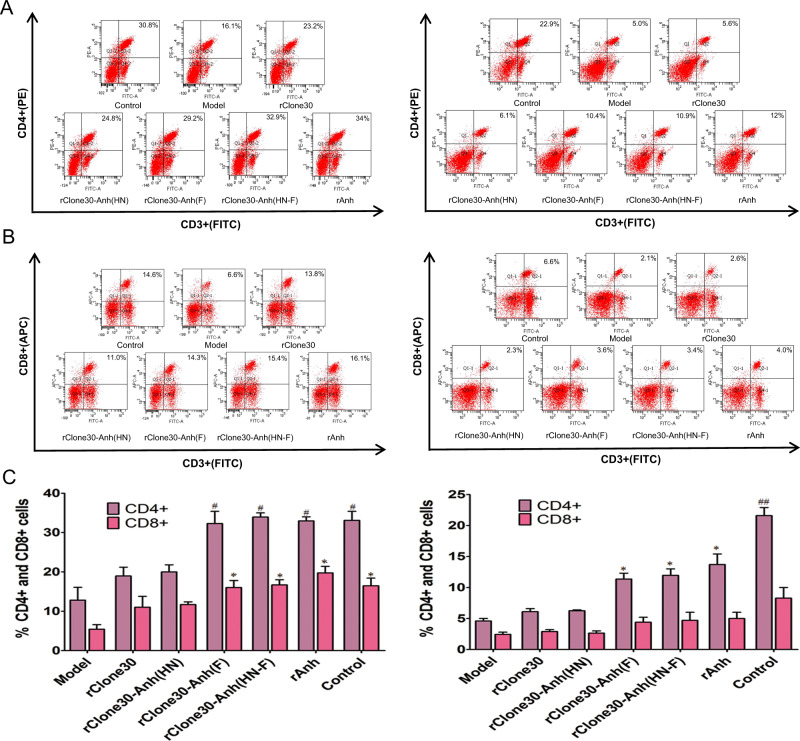
The immunomodulatory effect induced by recombinant viruses in BALB/c tumor-bearing mice was analyzed by flow cytometry. There was a significant decrease in the number of CD4+ and CD8+ T lymphocytes in the spleen of the tumor-bearing mice compared with control group in both H22 and 4T1-bearing mice (p < 0.05, Fig. 8). After treated with viruses, CD4+ and CD8+ T cells significantly increased, and more CD4+ and CD8+ T cells were detected in rClone30-Anh(F) and rClone30-Anh(HN-F)-infected groups than in rClone30 and rClone30-Anh(HN)-infected groups. There was no significant difference in the number of the T lymphocytes between rClone30-Anh(F) group and rClone30-Anh(HN-F) group.
Fig. 8. Chimeric viruses expressing the F gene of lytic strain Anhinga increase CD4+ T and CD8+ T cells.
a, b, c The CD4+ T and CD8+ T cells of the spleen in H22 (left) or 4T1 (right) tumor-bearing mice allantoic fluid-treated or treated with recombinant viruses were analyzed by flow cytometry, a CD4 + T cells. b CD8 + T cells. c The percentage of CD4+ T and CD8+ T cells. (Control: untreated mice. Model: mice treated with allantoic fluid. *P < 0.05, **P < 0.01, compared with the Model group. #P < 0.05, ##P < 0.01, n = 3, compared with rClone30 group).
rClone30-Anh(F) and rClone30-Anh(HN-F) demonstrate tumor specificity and safety
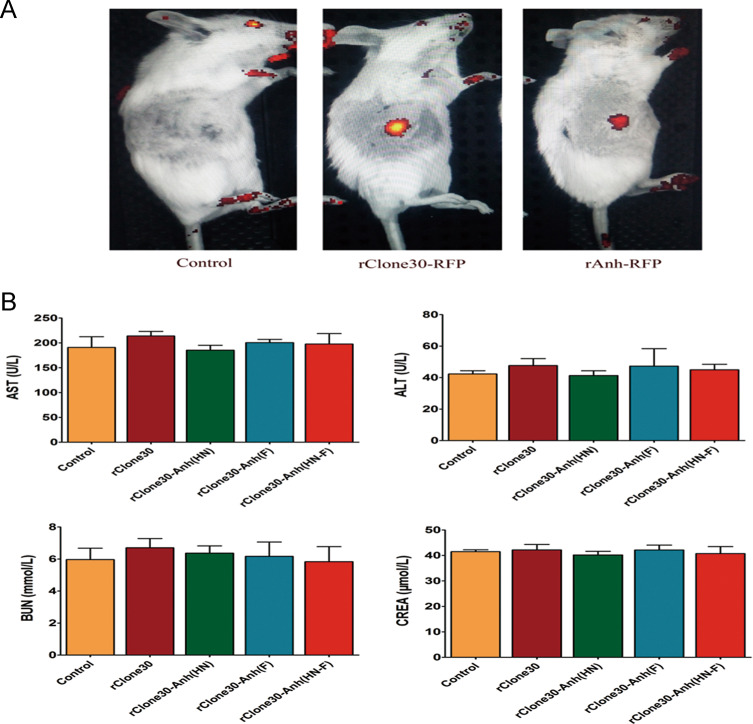
Recombinant viruses rClone30-RFP and rAnh-RFP were intravenously injected into subcutaneous tumor-bearing mice or normal mice to observe the distribution of viruses. As shown in Fig. 9a, the RFP was not detected in the tissues except tumor site, indicating that in one hand these viruses are tumor specific, in the other hand they are safe because virus is not distributed in the healthy tissues. After fifteen intraperitoneal injections of rClone30, rClone30-Anh(HN), rClone30-Anh(F), and rClone30-Anh(HN-F) for 30 days, the blood of normal mice and the mice injected with the viruses was collected. Serum chemistries revealed insignificant changes in BUN, creatinine levels, AST and ALT (Fig. 9b). These results suggest that NDV is safe for cancer therapy.
Fig. 9. NDV treatment demonstrates tumor specificity and safety.
a Recombinant viruses rClone30-RFP and rAnh-RFP was intravenously injected into subcutaneous tumor-bearing mice or normal mice to observe the distribution of virus. Control: normal mice intravenously treated with rAnh-RFP. rClone30-RFP: tumor-bearing mice intravenously injected with rClone30-RFP. rAnh-RFP: tumor-bearing mice intravenously injected with rAnh-RFP. b The serum biochemistry data of the experimental mice after injection of recombinant viruses. After 15 injections of rClone30, rClone30-Anh(HN), rClone30-Anh(F), and rClone30-Anh(HN-F), the blood of mice was drawn for measurement of serum chemistries. Control: normal mice. Values are expressed as mean ± SEM. ALT alanine aminotransferase, AST aspartate transaminase, BUN blood urea nitrogen, CREA creatinine.
Discussion
Since the first approval of the oncolytic virus, Amgen’s oncolytic herpes virus Imlygic (talimogene laherparepvec) in Europe and the United States, the OV field has gained much attraction. The therapeutic potential of oncolytic viruses is now being realized [33]. NDV as one of the oncolytic viruses represents a new class of therapeutic agents of cancer therapy. The oncolytic effect of wild and recombinant NDV strain has been demonstrated for being effective by multiple studies in experimental models and clinical trials [34–36]. Though the great progress in the oncolytic effect of NDV has been made by humanity, there still remain many challenges.
One of the major challenges of oncolytic virotherapy is that the susceptibility to oncolysis ranges among different cancer cell types. Li [37] had reported that the replication of M1 highly associates with oncolysis. The large-scale screen of cell line revealed that the susceptibility to M1 are quite variable, cells infected with M1 oncolytic virus could be defined as hypersensitive cell, sensitive cell, and refractory cell. M1 replication was significantly higher in hypersensitive cancer cells and lower in refractory cancer cells, indicating that it is important for enhancing the oncolytic effect to investigate the susceptibility of cancer cells to oncolytic virus infection. Previous study showed that a broad spectrum of human cancer cells including tumor cell lines of ecto-, endo-, and meso-dermal origin [37] were killed by NDV in vitro, but our unpublished data showed the susceptibility of cancer cell lines to NDV infection was substantially different. It is necessary to conduct a systematic study on the susceptibility of cancer cells to NDV infection. According to global cancer statistics 2018, the incidence or mortality of lung cancer, breast cancer, colorectal cancer, liver cancer, and brain cancer are high [38]. Among the five types of cancer, we chose the representative cell lines HepG2, MDA-MB-231, SH-SY5Y, A549, MCF7, and LoVo to investigate the susceptibility of cancer cells to NDV infection. The susceptibility of cancer cells to non-lytic strain (rClone30-RFP) and lytic strain (rAnh-RFP) was determined by the expression of RFP, the number of syncytia formation, and the inhibition rate of the two viruses on cancer cells. We found that the susceptibility of cancer cells to NDV infection is different and the infection of NDV are higher in HepG2, MDA-MB-231, SH-SY5Y cells (the fluorescent intensity were 74.9%, 52.25%, and 44.3%) (hypersensitive cancer cells), intermediate in A549, and MCF7 cells (the fluorescent intensity were 27.65% and 33.5%) (sensitive cancer cells), lower in LoVo cells (the fluorescent intensity were 6.7%) (refractory cancer cells). The cytotoxicity of rClone30-RFP, rAnh-RFP showed the same trends, indicating that the infection and replication ability of NDV is positively correlated with the effect of oncolytic effect.
In the six cells involved in viral infection experiments, only MDA-MB-231 and MCF7 derive from the same tissue and thus have more similar genetic background, contributing to precisely determined which gene is responsible for the susceptibility to NDV infection. To investigate the molecular mechanism which is responsible for cancer cell susceptibility to NDV infection, transcriptome sequencing was carried out to analyze the differentially expressed genes between susceptible MDA-MB-231 and less susceptible MCF7. Cells without viral infection served as a negative control. The results showed that many genes were differentially expressed after MDA-MB-231 and MCF7 cells infected with viruses (Fig. 3a). Here, we focus on sialic acid and IFN. Previous research showed the abundant presence of sialoglycoproteins on the surface of cancer cells most likely promotes preferential association of the virus with malignant and contributes to their selective oncolytic effect [39, 40]. Furthermore, it was confirmed that α2,6-linked sialic acid serves as a high-affinity receptor for NDV therapy. ST6GalI-transfected CHO-K1 cells increase the α2,6-linkage level, enhancing NDV binding and cytopathic effect [41]. We found the transcriptional level of alpha-sialic acid acyltransferase (ST6GalI) highly expressed in MDA-MB-231 cells than that in MCF7 cells (Fig. 3b). Meanwhile, previous studies show that the replication and spread of NDV is significantly reduced in normal cells compared with cancer cells due to antiviral signaling pathways [9]. After cancer cells were infected with NDV, the viral replication can be promoted specifically in tumors due to the specific genetic defects of the IFN and apoptotic pathways based on type I IFN signaling [42]. IRF3 and IRF7 are the main regulators of type I IFN expression, when cells are stimulated with virus infection, IRF3 are phosphorylated by the serine/threonine kinases, TBK1 and IKKɛ, and then homodimerized IRF3 translocates from the cytosol into the nucleus and binds to responsive elements for IFN-β gene transcription [43]. GLTSCR2 as a nuclear protein support the viral replication of paramyxovirus in cells. Viral infection induced translocation of GLTSCR2 from nucleus to cytoplasm that enabled GLTSCR2 to attenuate type I interferon IFN-β and promote viral replication [44]. In this study, when the changes of these genes mentioned above in two kinds of cells (susceptible MDA-MB-231 and less susceptible MCF7) infected with NDV were analyzed, we observed that, compared with MCF7 cells, the expression of GLTSCR2 were upregulated in MDA-MB-231 and the expression of IFN-related genes was downregulated. We further identified that, though cancer cell are specifically infected by NDV, the cancer cell susceptibility to NDV infection was different, sialic acid and the IFN were responsible for cancer cell susceptibility. These genes can be used as markers of cancer cell susceptibility to NDV infection, which lays a foundation for clinical screening of susceptible tumor types. But the most important genes for cell susceptibility to viruses infection need more study.
To study the effect of cancer cell susceptibility on the NDV treatment, the oncolytic effects of chimeric virus on susceptible cell lines HepG2, MDA-MB-231, H22 and less susceptible cell lines A549, MCF7, and 4T1 were tested (Fig. 2 and Supplementary Fig. 2A). In vitro, compared with less susceptible cell lines, a huge syncytium was induced by rClone30-Anh(F) and rClone30-Anh(HN-F) on susceptible cell lines (Fig. 4a and Supplementary Fig. 2B). The growth inhibition of rClone30-Anh (F) and rClone30-Anh (HN-F) on susceptible cells was stronger than that on less susceptible cells (greater than twofold change) (Fig. 4b and Supplementary Fig. 2C). When the cells were infected with virus of 0.1 MOI for 24 h, the apoptosis rate induced by rClone30-Anh(F) in HepG2 cells was 15.55% and 6.75% in MCF7 cells (Fig. 4c, d). In vivo experiments, to investigate the oncolytic effect of the chimeric viruses on xenographic mice derived from NDV susceptible (H22) and less susceptible (4T1) cells, the xenographic mice were intratumorally injected with allantoic fluid or 1 × 108 pfu of the indicated virus every day. In H22 tumor-bearing mice, rClone30-Anh(F) and rClone30-Anh(HN-F) showed a more obvious suppression in tumor volume, tumor weight, and tumor apoptosis rate than that in 4T1 tumor-bearing mice (Fig. 6). In vivo and in vitro experiments were consistent, rClone30-Anh(F) and rClone30-Anh(HN-F) showed a more obvious oncolytic effect in the susceptible cells, suggesting that NDV play a more effective antitumor role in susceptible cells.
It is reported that NDV was classified by their pathogenesis in birds as lentogenic (avirulent, no clinical manifestations), mesogenic (intermediate, mild disease), or velogenic (virulent, high mortality) and none of the strains are associated with serious disease in humans [45]. All NDV strains are associated with antitumor immune activity, the mesogenic and velogenic types demonstrate a lytic replication cycle in human tumor cells leading to an amplification of the viral load but non-lytic lentogenic strains not, which reveals that lytic strains are more effective than non-lytic strains in tumor cytotoxicity. Normally, a lentogenic strain was chosen as an antitumor agent because of the minimal hazard for environment but the oncolytic effect of NDV will be limited [45]. The aim of research is to produce optimized NDV Clone30 vectors via virus-engineering strategies, which not only provide more potent cytotoxic effects, but also demonstrate enhanced safety profiles [46]. Considering the positive and negative features of both lytic strains (mesogenic, rAnh) and non-lytic strains (lentogenic, rClone30) as oncolytic virus platforms, we engineered a chimeric virus based on the lentogenic NDV backbone, in which the oncolytic ability was altered.
Extensive research in F gene has been performed to elucidate the molecular determinants for viral pathogenicity and fusion activity [47, 48], showing the F protein is the major contributor for virulence, and F protein is responsible for viral fusion with formation of syncytia [49]. The HN gene also plays an important role in NDV infection and budding process. Previous studies have shown that the stalk region of HN protein is also implicated in the activation of the fusion activity of the F protein [50]. Estevez et al. showed that the mutation at position 192 (Ile-Met) of HN proteins derived from the mesogenic NDV strain Anhinga decreased haemadsorption and neuraminidase activities and increased fusion promotion activity [51]. In addition, during viral infection, the binding of the HN protein to its cognate receptor, cellular sialic acid, causes a conformational change that facilitates the specific homotypic interaction between F and HN, triggering fusion of the viral envelope with the host cell membrane [52] indicating that the interaction of homotypic HN and F proteins of paramyxoviruses is important for the proper function of these proteins.
To clarify which protein (F or HN) is mainly responsible for the oncolytic capacity of the NDV, a series of chimeric viruses rClone30-Anh(HN), rClone30-Anh(F), and rClone30-Anh(HN-F) were constructed by exchanging the HN gene, F gene, or both of non-lytic rClone30 strain with lytic strain Anhinga. In vitro, little syncytia was induced by rClone30 and rClone30-Anh(HN), but a huge syncytia was induced by rClone30-Anh(F) and rClone30-Anh(HN-F). The cytotoxicity of rClone30-Anh(F), rClone30-Anh(HN-F) to cancer cells was significant higher than that of rClone30 and rClone30-Anh(HN) (p < 0.05, Fig. 4). In vivo, in comparison with rClone30 and rClone30-Anh(HN), the tumor volume of H22 and 4T1 tumor-bearing mice was significantly inhibited by rClone30-Anh(F) and rClone30-Anh(HN-F), the 100-day survival rate of rClone30-Anh(F), rClone30-Anh(HN-F)-treated mice was increased by 40% compared with rClone30, rClone30-Anh(HN)-treated mice in H22-bearing model, while 20% in 4T1-bearing model (Fig. 6). HE and TUNEL analysis showed an increased tumor apoptosis rate in rClone30-Anh(F), rClone30-Anh(HN-F)-treated mice compared with that in rClone30, rClone30-Anh(HN)-treated mice (Fig. 7). The more CD4+ T and CD8+ T-cell populations were found in the splenocytes isolated from the tumor-bearing mice treated with rClone30-Anh(F) or rClone30-Anh(HN-F) and the unpublished data showed rClone30-Anh(F) or rClone30-Anh(HN-F) treatment downregulated the immunosuppressive factor, which suggest that chimeric viruses expressing the F gene of lytic strain Anhinga can circumvent immune suppression and enhance the systemic antitumour immunity (Fig. 8). Meanwhile, both rClone30-Anh(F) and rClone30-Anh(HN-F) showed a decreased pathogenicity compared with parental virus Anhinga (lower ICPI score and higher MDT) (Table 2). These results indicate that the F protein plays a major role in NDV-induced oncolytic effect on xenograft derived from cancer cells (susceptible cell lines H22 and less susceptible cell lines 4T1). The F gene of lentogenic rClone30 with clear genetic background is recommended to be exchanged with the F gene of NDV strains with strong fusion ability to enhance the oncolytic effect of the wild-type rClone30.
To elucidate the oncolytic mechanism of rClone30-Anh(F), the differential gene expression of MDA-MB-231 cells infected with rClone30 or rClone30-Anh(F) was analyzed by transcriptome sequencing (Supplementary Fig. 1). Previous studies reported that the LC3/LC3II turnover assay is used as a classical method to investigate the autophagic flux dynamics [53]. Autophagy related 5 (ATG5) as a key protein in the extension of the phagophoric membrane in autophagic vesicles is necessary for LC3I conjugation to phosphatidylethanolamine to form LC3II [54]. The other autophagy marker Becline-1 positively regulates autophagy by generating membrane-bound protein complexes, promoting the formation of autophagosome [55]. As shown in Fig. 5a, b, transcriptome analysis and real-time PCR analysis revealed that the genes ATG5, becline-1, and MAP1LC3B were upregulated in MDA-MB-231 cells infected with rClone30-Anh(F). Our data has shown cell death induced by rClone30-Anh(F) and rClone30 was different, and maybe the oncolytic mechanism of rClone30-Anh(F) is associate with autophagy but rClone30 not. Ren et al. [24] show that the syncytia is generated by activating AMPK-mTORC1-ULK1 signaling. Under nutrient-rich conditions, the suppression of autophagy can be leaded since mTORC1 phosphorylates unc-51 like autophagy activating kinase 1 (ULK1) inhibit its interaction with AMPK [56]. But, cellular energy depletion induces AMPK-mediated mTORC1 inhibition and then leads to autophagy induction [54]. Ragulator as a key protein complex in the mTOR signaling pathway plays an important role in binding Rag GTPases to the surface of lysosomes in the mTOR pathway and activating mTORC1. mTORC1 is a complex composed of p18, p14, MP1, Lamtor4, and Lamtor5 proteins encoded by LAMTOR1, LAMTOR2, LAMTOR3, C7orf59, and HBXI [57, 58]. The results showed that, in comparison with rClone30, rClone30-Anh(F) downregulated the transcriptional level of LAMTOR1, LAMTOR2, LAMTOR4, LAMTOR5, and RHEB GTP-binding protein, but upregulated the transcriptional level of ULK1 (Fig. 5a). We made a hypothesis that mTORC1 is inhibited by rClone30-Anh(F), thus leading to autophagy induction, but there still are much work need to be done to confirm that rClone30-Anh(F) activates AMPK-mTOR-ULK1 signaling for the initiation of a complete autophagic flux and the subsequent formation of syncytia.
In conclusion, this study elaborated the methods to improve the oncolytic effect of NDV Clone30 from two aspects of host cells and virus per se. The results showed that the susceptibility of cancer cells to NDV infection was substantially different and NDV treatment is more effective in susceptible cells, which was related to the expression of alpha-sialic acid acyltransferase and interferon. rClone30-Anh(F) and rClone30-Anh(HN-F) as oncolytic agents showed a stronger oncolytic effect compared with rClone30, indicating that the F gene plays an important role in oncolytic capability of NDV in vivo. Thereby we offered an ideal strategy to enhance the oncolytic effect of NDV Clone30 by the selecting of susceptible tumor type as indications and constructing chimeric virus based on lentogenic virus with F gene derived from virus with strong fusion ability in consideration to the epidemiology of NDV in a giving region.
Supplementary information
Supplementary information :Figure-1 and Figure-2
Genes regulated by rClone30 or rClone30-Anh(F) was different.
Mouse hepatocarcinoma cell H22 is more susceptible than mouse breast cancer cell 4T-1.
Acknowledgements
This work was financially supported by National Key R&D Program of China (2017YFD0501102, 2017YFD0501103-03, and 2017YFD0501004), National Natural Science Foundation of China (31600740) and the Jiangsu Provincial Key Laboratory for Bioresources of saline soils (JKLBS2017010).
Author contributions
CRediT authorship contribution statement. TL: conceptualization, methodology, validation, formal analysis, investigation, writing-original draft, and visualization. DL: conceptualization, methodology, validation, supervision, resources, and data curation. WX: methodology, project administration, funding acquisition, and data curation. YZ: formal analysis, investigation, validation, and software. YC: validation and investigation. SJ: validation and software. RS: formal analysis and investigation. JY: methodology and conceptualization. ZG: resources and funding acquisition. ZW: project administration and funding acquisition. GR: methodology and resources. QY: investigation and resources. GS: methodology and resources. XS: formal analysis and investigation. WS: investigation.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1038/s41434-024-00459-9
Change history
7/4/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41434-024-00459-9
Contributor Information
Wei Xiao, Email: xw_kanion@163.com.
Deshan Li, Email: deshanli@163.com.
Supplementary information
The online version of this article (10.1038/s41434-020-0145-9) contains supplementary material, which is available to authorized users.
References
- 1.Schirrmacher V. Clinical trials of antitumor vaccination with an autologous tumor cell vaccine modified by virus infection: improvement of patient survival based on improved antitumor immune memory. Cancer Immunol Immunother. 2005;54:587–98. doi: 10.1007/s00262-004-0602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman AI, Zakay-Rones Z, Gomori JM, Linetsky E, Rasooly L, Greenbaum E, et al. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol Ther. 2006;13:221–8. doi: 10.1016/j.ymthe.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Schirrmacher V, Fournier P. Newcastle disease virus: a promising vector for viral therapy, immune therapy, and gene therapy of cancer. Methods Mol Biol. 2009;542:565–605. doi: 10.1007/978-1-59745-561-9_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schirrmacher V. Fifty years of clinical application of newcastle disease virus: time to celebrate! Biomedicines. 2016;4:16. doi: 10.3390/biomedicines4030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schirrmacher V, van Gool S, Stuecker W. Breaking therapy resistance: an update on oncolytic newcastle disease virus for improvements of cancer therapy. Biomedicines. 2019;7:66. doi: 10.3390/biomedicines7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matveeva OV, Zong SG, Shabalina SA, Chumakov PM. Oncolysis by paramyxoviruses: multiple mechanisms contribute to therapeutic efficiency. Mol Ther Oncolytics. 2015;2:15011.. doi: 10.1038/mto.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cattaneo R, Miest T, Shashkova E, Barry M. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat Rev Microbiol. 2008;6:529–40. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilan Y, Sauter B, Chowdhury NR, Reddy BV, Thummala NR, Droguett G, et al. Oral tolerization to adenoviral proteins permits repeated adenovirus-mediated gene therapy in rats with pre-existing immunity to adenoviruses. Hepatology. 1998;27:1368–76. doi: 10.1002/hep.510270525. [DOI] [PubMed] [Google Scholar]
- 9.Dmitriy Z, Peter P. Oncolytic Newcastle disease virus for cancer therapy: old challenges and new directions. Future Microbiol. 2012;7:347–67. doi: 10.2217/fmb.12.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christoph F, Ben P, Philippe F, Annette A, Mariana B, Volker S. Tumor selective replication of Newcastle disease virus: association with defects of tumor cells in antiviral defence. Int J Cancer. 2010;119:328–38. doi: 10.1002/ijc.21821. [DOI] [PubMed] [Google Scholar]
- 11.Zamarin D, Martinezsobrido L, Kelly K, Mansour M, Sheng G, Vigil A, et al. Enhancement of oncolytic properties of recombinant newcastle disease virus through antagonism of cellular innate immune responses. Mol Ther. 2009;17:697–706. doi: 10.1038/mt.2008.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu Z, Bai F, Sun T, Tian H, Yu D, Yin J, et al. Recombinant newcastle disease virus expressing IL15 demonstrates promising antitumor efficiency in melanoma model. Technol Cancer Res Treat. 2014;14:607–15. doi: 10.7785/tcrt.2012.500414. [DOI] [PubMed] [Google Scholar]
- 13.Yang B, Chen Y, Hong X, Liu X, Su X, Li S, et al. Newcastle disease virus enhances the growth-inhibiting and proapoptotic effects of temozolomide on glioblastoma cells in vitro and in vivo. Sci Rep. 2018;8:11470-. doi: 10.1038/s41598-018-29929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2016;15:660. doi: 10.1038/nrd.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Pan Z, Tian G, Xin L, Liu Y, Guo X, et al. Newcastle disease virus chimeras expressing the Hemagglutinin- Neuraminidase protein of mesogenic strain exhibits an enhanced anti-hepatoma efficacy. Virus Res. 2016;221:23–29. doi: 10.1016/j.virusres.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Welch BD, Marcin P, Leser GP, Zachary B, Kors CA, Paterson RG, et al. Probing the functions of the paramyxovirus glycoproteins F and HN with a panel of synthetic antibodies. J Virol. 2014;88:11713–25. doi: 10.1128/JVI.01707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji Y, Liu T, Jia Y, Liu B, Yu Q, Cui X, et al. Two single mutations in the fusion protein of Newcastle disease virus confer hemagglutinin-neuraminidase independent fusion promotion and attenuate the pathogenicity in chickens. Virology. 2017;509:146–51. doi: 10.1016/j.virol.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Pingdong L, Chun-Hao C, Sen L, Babak G, Zhenkun Y, Dmitriy Z, et al. Therapeutic effects of a fusogenic newcastle disease virus in treating head and neck cancer. Head Neck. 2011;33:1394–9. doi: 10.1002/hed.21609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Bi Y, Yu W, Wei N, Wang W, Wei Q, et al. Two mutations in the HR2 region of Newcastle disease virus fusion protein with a cleavage motif “RRQRRL” are critical for fusogenic activity. Virol J. 2017;14:185. doi: 10.1186/s12985-017-0851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chengxi S, Hongling W, Yuzhen C, Fulu C, Bin L, Guijie R, et al. Roles of the highly conserved amino acids in the globular head and stalk region of the Newcastle disease virus HN protein in the membrane fusion process. Biosci Trends. 2015;9:56. doi: 10.5582/bst.2014.01140. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Quinlan EA, Iorio RM. Mutated form of the Newcastle disease virus hemagglutinin-neuraminidase interacts with the homologous fusion protein despite deficiencies in both receptor recognition and fusion promotion. J Virol. 2004;78:5299–310. doi: 10.1128/JVI.78.10.5299-5310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangaswamy US, Wang W, Cheng X, McTamney P, Carroll D, Jin H, et al. Newcastle disease virus establishes persistent infection in tumor cells in vitro: contribution of the cleavage site of fusion protein and second sialic acid binding site of hemagglutinin-neuraminidase. J Virol. 2017;91:e00770–17. doi: 10.1128/JVI.00770-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsurudome M, Ohtsuka J, Ito M, Nishio M, Nosaka T. The hemagglutinin-neuraminidase (HN) head domain and the fusion (F) protein stalk domain of the parainfluenza viruses affect the specificity of the HN-F Interaction. Front Microbiol. 2018;9:391. doi: 10.3389/fmicb.2018.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren S, Rehman ZU, Shi M, Yang B, Qu Y, Yang XF, et al. Syncytia generated by hemagglutinin-neuraminidase and fusion proteins of virulent Newcastle disease virus induce complete autophagy by activating AMPK-mTORC1-ULK1 signaling>. Vet Microbiol. 2019;230:283–90. doi: 10.1016/j.vetmic.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Ren S, Rehman ZU, Shi M, Yang B, Liu P, Yin Y, et al. Hemagglutinin-neuraminidase and fusion proteins of virulent Newcastle disease virus cooperatively disturb fusion-fission homeostasis to enhance mitochondrial function by activating the unfolded protein response of endoplasmic reticulum and mitochondrial stress. Vet Res. 2019;50:37. doi: 10.1186/s13567-019-0654-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Yan S, Lv Z, Chen L, Geng J, He J, et al. Recombinant Newcastle disease virus Anhinga strain (NDV/Anh-EGFP) for hepatoma therapy. Technol Cancer Res Treat. 2014;13:169–75. doi: 10.7785/tcrt.2012.500356. [DOI] [PubMed] [Google Scholar]
- 27.Bai F, Niu Z, Tian H, Li S, Lv Z, Zhang T, et al. Genetically engineered Newcastle disease virus expressing interleukin 2 is a potential drug candidate for cancer immunotherapy. Immunol Lett. 2014;159:36–46. doi: 10.1016/j.imlet.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–7. [Google Scholar]
- 29.Alexander DJ. Newcastle disease and other avian paramyxoviruses. Rev Sci Tech. 2000;19:443. doi: 10.20506/rst.19.2.1231. [DOI] [PubMed] [Google Scholar]
- 30.Pan Z, He J, Rasoul LM, Liu Y, Che R, Ding Y, et al. Identification of optimal insertion site in recombinant newcastle disease virus (rNDV) vector expressing foreign gene to enhance its anti-tumor effect. PLoS ONE. 2016;11:e0164723. doi: 10.1371/journal.pone.0164723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Ayeka PA, Bian YH, Githaiga PM, Zhao Y. The immunomodulatory activities of licorice polysaccharides (Glycyrrhiza uralensis Fisch.) in CT26 tumor-bearing mice. BMC Complement Altern Med. 2017;17:536. doi: 10.1186/s12906-017-2030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffy MR, Fisher KD, Seymour LW. Making oncolytic virotherapy a clinical reality: the European contribution. Human Gene Ther. 2017;28:1033–46. doi: 10.1089/hum.2017.112. [DOI] [PubMed] [Google Scholar]
- 34.Volker S, Christoph S, Jurgen W, Philipp B. Strong T-cell costimulation can reactivate tumor antigen-specific T cells in late stage metastasized colorectal carcinoma patients: results from a phaseI clinical study. Int J Oncol. 2015;46:71–7. doi: 10.3892/ijo.2014.2692. [DOI] [PubMed] [Google Scholar]
- 35.Marianna P, Judit B, Jozsef S. Sensitivity of human malignant melanoma cell lines to Newcastle disease virus. Anticancer Res. 2015;35:5401–6. [PubMed] [Google Scholar]
- 36.Ahmad U, Ahmed I, Keong YY, Abd MN, Othman F. Inhibitory and apoptosis-inducing effects of Newcastle disease virus strain AF2240 on mammary carcinoma cell line. Biomed Res Int. 2015;2015:127828. doi: 10.1155/2015/127828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li K, Zhang H, Qiu J, Lin Y, Liang J, Xiao X, et al. Activation of cyclic adenosine monophosphate pathway increases the sensitivity of cancer cells to the oncolytic virus M1. Mol Ther. 2016;24:156–65. doi: 10.1038/mt.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 39.Rui X, Palmer SG, Matteo P, Palermo LM, Stefan N, Wilson IA, et al. Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo. mBio. 2013;4:00803–13. doi: 10.1128/mBio.00803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bose S, Jardetzky TS, Lamb RA. Timing is everything: fine-tuned molecular machines orchestrate paramyxovirus entry. Virology. 2015;479–80:518–31. doi: 10.1016/j.virol.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Wei D, Feng F, Wang XL, Li C, Chen ZN, et al. alpha2,6-linked sialic acid serves as a high-affinity receptor for cancer oncolytic virotherapy with Newcastle disease virus. J Cancer Res Clin Oncol. 2017;143:2171–81. doi: 10.1007/s00432-017-2470-y. [DOI] [PubMed] [Google Scholar]
- 42.Matveeva OV, Guo ZS, Shabalina SA, Chumakov PM. Oncolysis by paramyxoviruses: multiple mechanisms contribute to therapeutic efficiency. Mol Ther Oncolytics. 2015;2:15011. doi: 10.1038/mto.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilden H, Fournier P, Zawatzky R, Schirrmacher V. Expression of RIG-I, IRF3, IFN-beta and IRF7 determines resistance or susceptibility of cells to infection by Newcastle Disease Virus. Int J Oncol. 2009;34:971–82. doi: 10.3892/ijo_00000223. [DOI] [PubMed] [Google Scholar]
- 44.Wang P, Meng W, Han S-C, Li C-C, Wang X-J, Wang X-J. The nucleolar protein GLTSCR2 is required for efficient viral replication. Sci Rep. 2016;6:36226. doi: 10.1038/srep36226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam HY, Yeap SK, Pirozyan MR, Omar AR, Yusoff K, Abd-Aziz S, et al. Corrigendum to “Safety and Clinical Usage of Newcastle Disease Virus in Cancer Therapy”. Biomed Res Int. 2017;2017:4529437. doi: 10.1155/2017/4529437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdullahi S, Jäkel M, Behrend SJ, Steiger K, Topping G, Krabbe T, et al. A novel chimeric oncolytic virus vector for improved safety and efficacy as a platform for the treatment of hepatocellular carcinoma. J Virol. 2018;92:e01386–18. doi: 10.1128/JVI.01386-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kai Y, Hu Z, Xu H, Hu S, Zhu J, Hu J, et al. The M, F and HN genes of genotype VIId Newcastle disease virus are associated with the severe pathological changes in the spleen of chickens. Virol J. 2015;12:133. doi: 10.1186/s12985-015-0366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakamatsu N, King DJ, Seal BS, Samal SK, Brown CC. The pathogenesis of Newcastle disease: a comparison of selected Newcastle disease virus wild-type strains and their infectious clones. Virology. 2006;353:333–43. doi: 10.1016/j.virol.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 49.Paldurai A, Kim SH, Nayak B, Xiao S, Shive H, Collins PL, et al. Evaluation of the contributions of individual viral genes to newcastle disease virus virulence and pathogenesis. J Virol. 2014;88:8579–96. doi: 10.1128/JVI.00666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–15. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 51.Estevez C, King DJ, Luo M, Yu Q. A single amino acid substitution in the haemagglutinin-neuraminidase protein of Newcastle disease virus results in increased fusion promotion and decreased neuraminidase activities without changes in virus pathotype. J Gen Virol. 2010;92:544–51. doi: 10.1099/vir.0.027540-0. [DOI] [PubMed] [Google Scholar]
- 52.Tsurudome M, Ito M, Nishio M, Nakahashi M, Kawano M, Komada H, et al. Identification of domains on the fusion (F) protein trimer that influence the hemagglutinin-neuraminidase specificity of the f protein in mediating cell-cell fusion. J Virol. 2011;85:3153–61. doi: 10.1128/JVI.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang Y, Yuan R, Xiang B, Zhao X, Gao P, Dai X, et al. Newcastle disease virus-induced autophagy mediates antiapoptotic signaling responses in vitro and in vivo. Oncotarget. 2017;8:73981–93. doi: 10.18632/oncotarget.18169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klionsky DJ, Abdalla FC, Hagai A, Abraham RT, Abraham AA, Khosrow A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen S, Li L, Li S, Bai Y, Liu H. Metal-organic frameworks induce autophagy in mouse embryonic fibroblast cells. Nanoscale. 2018;10. 10.1039/c8nr04459g. [DOI] [PubMed]
- 56.Joungmok K, Mondira K, Benoit V, Kun-Liang G. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Araujo ME, Stasyk T, Taub N, Ebner HL, Furst B, Filipek P, et al. Stability of the endosomal scaffold protein LAMTOR3 depends on heterodimer assembly and proteasomal degradation. J Biol Chem. 2013;288:18228–42. doi: 10.1074/jbc.M112.349480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information :Figure-1 and Figure-2
Genes regulated by rClone30 or rClone30-Anh(F) was different.
Mouse hepatocarcinoma cell H22 is more susceptible than mouse breast cancer cell 4T-1.