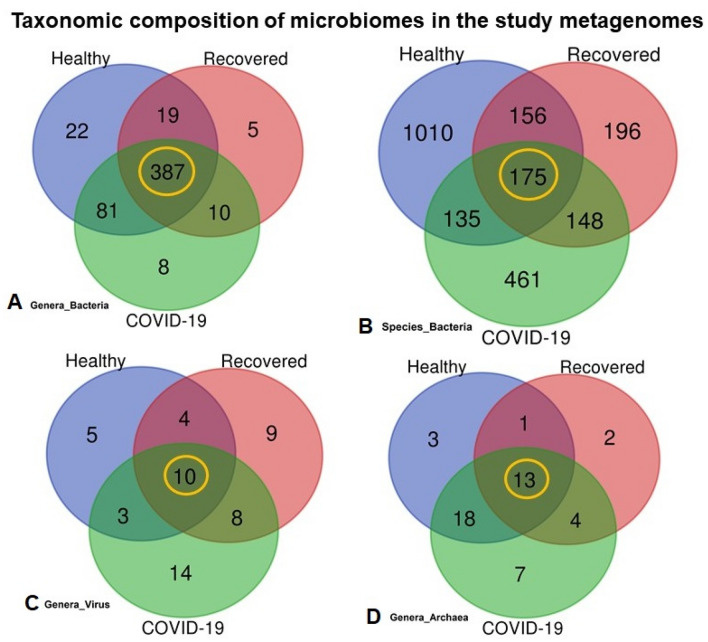
Figure 3.
Taxonomic composition of the nasopharyngeal microbiomes of COVID-19 patients, Recovered people and Healthy controls. Venn diagrams representing the core unique and shared microbiomes in COVID-19, Recovered and Healthy samples. (A) Venn diagram showing unique and shared bacterial genera. Out of 532 genera detected, only 387 genera (highlighted in yellow) were found to be shared across three sample categories. (B) Venn diagram comparison of unique and shared bacterial species where only 175 (highlighted in yellow) species shared in the study metagenomes, and 461, 196 and 1,010 species had sole association with COVID-19, Recovered and Healthy samples, respectively. (C) Venn diagrams representing unique and shared viral genera identified in three metagenomes. Of the detected viral genera (n = 53), 14, 9 and 7 genera had unique association with COVID-19, Recovered and Healthy samples, respectively, and 10 genera (highlighted in yellow) were found to be shared across three metagenomes. (D) Venn diagrams showing unique and shared archaeal genera in COVID-19, Recovered and Healthy nasopharyngeal samples. A total of forty-eight archaeal genera (n = 48) was detected, of which 7, 2 and 3 genera had unique association with COVID-19, Recovered and Healthy samples, respectively, and 13 genera (highlighted in yellow) were found to be shared across three metagenomes. More information on the taxonomic result is also available in Data S1. Venn diagrams are generated through an online tool used for Bioinformatics and Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/).