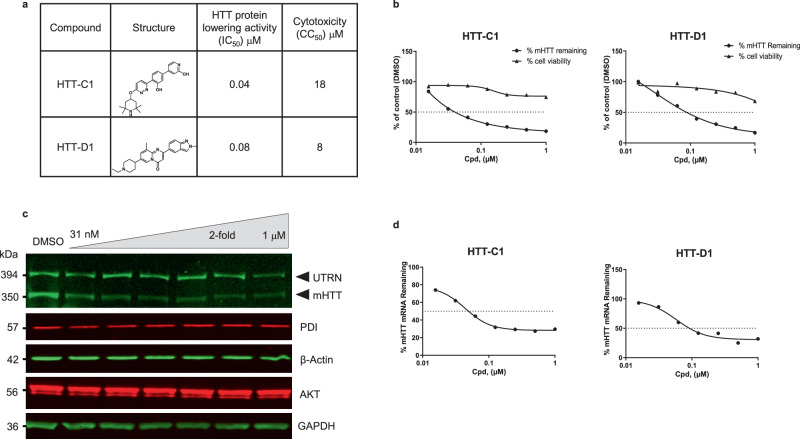
Fig. 1. Huntingtin (HTT)-lowering activity in vitro.
a Chemical structures of HTT-C1 and HTT-D1. b Electrochemiluminescence (ECL) analysis of mutant HTT protein from fibroblasts isolated from a homozygous patient with Huntington’s disease (HD) (GM04857) after 96 h of continuous treatment with HTT-C1 and HTT-D1 (0.01–1.0 μM). Representative graphs show percent HTT remaining relative to the dimethyl sulphoxide (DMSO) control. Cell viability assays were performed in parallel. Data represent mean of two (n = 2) biologically independent samples per data point from one dose–response experiment. c Western blot of HTT protein and housekeeping proteins, oxidoreductase-protein disulphide isomerase (PDI), beta-actin, alpha serine/threonine-protein kinase (AKT) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in HD fibroblasts after 96 h of continuous treatment with HTT-C1 (0.015–1.0 μM). Utrophin (UTRN) was also used as a loading control. The western blot data used a representative splicing modifier (tested at multiple concentrations) from a single experiment. Multiple splicing modifiers from the same class were tested and evaluated by western blot analyses. d Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of HTT mRNA in patient fibroblasts after 24 h of treatment with HTT-C1 and HTT-D1 (0.01–1.0 μM). Representative graphs show percent HTT mRNA remaining relative to DMSO control; normalised to housekeeping gene, TATA-box binding protein. Data represent mean of two (n = 2) biologically independent samples per data point from one dose–response experiment.