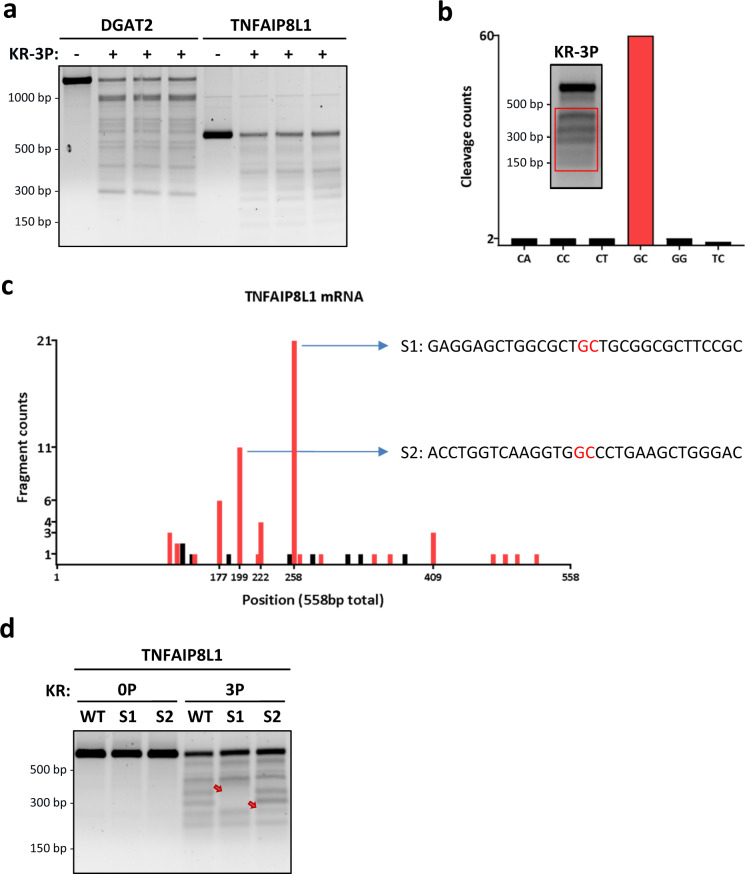
Fig. 3. RIDDLE is more promiscuous in substrate recognition, yet non-random.
a Comparative KR-3P digestion of DGAT2 and TNFAIP8L1 transcripts performed in three independent experiments. b Amount of RNA fragments sequenced whose 3′ end leads to the cleaved nt pair designated on the x axis. The first nt in the pair represents the last sequenced nt from the RNA fragment, while the second shows the subsequent base in the RNA sequence. Inset: red box indicates the portion of the gel that was extracted for Sanger sequencing. c Mapping of the last base pair (3′ end) from each individual RNA fragment sequenced within the TNFAIP8L1 mRNA. Red bars indicate cleavage sites between a GC nt pair. Black bars indicate non GC cleavage sites. d RNA digestions of WT TNFAIP8L1 and TNFAIP8L1 mutated at locations S1 and S2. The red arrows indicate a change in banding pattern as compared to WT.