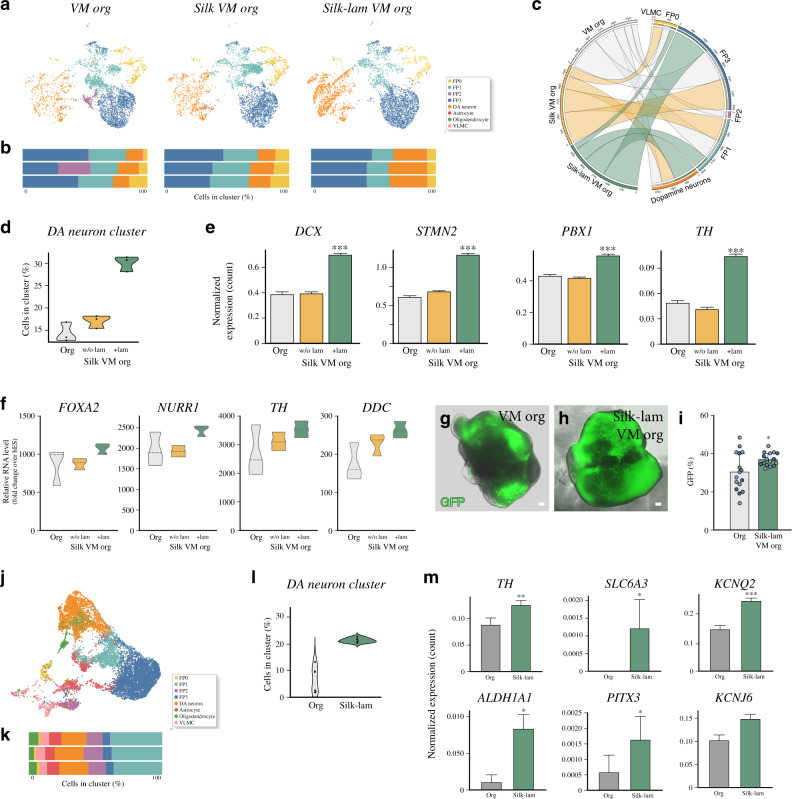
Fig. 6. Single-cell transcriptomics identifying silk-VM organoid cell composition.
a UMAP plots showing cell clusters from conventional VM, silk-VM, and silk-lam VM organoids and b, percentage of cells belonging to each cell cluster from individual organoids at month 1. c Chord diagram visualizing cell-type interrelationships between conventional, silk-VM and silk-lam VM organoids. d Violin plot showing the percentage of cells belonging to DA neuron clusters from conventional, silk-VM, and silk-lam VM organoids at month 1 from three individual organoids per condition. e Expression of selected markers belonging to DA neuron cluster in conventional, silk-VM, and silk-lam VM organoids at 1 month. Data represent mean ± SEM of 3 biologically independent organoids, two-tailed Wilcoxon Rank Sum test, ***p < 0.0001. f qRT-PCR analysis of early and late DA neuron markers in conventional and silk/silk-lam VM individual organoids at month 2. Data represent mean ± SEM of 3 independent organoids per condition. g, h Representative images of GFP expression in conventional and silk-lam VM organoids differentiated from the CRISPR/Cas9-mediated gene-edited TH-Cre hPSC line. Scale bars, 100 µm. i FACS-based quantification of GFP expression in conventional and silk-lam VM organoids differentiated from a CRISPR/Cas9-mediated gene-edited TH-Cre hPSC line in 4 biologically independent experiments shown as color-coded dots (green, light blue, blue and purple). Data represent mean ± SD, two-tailed unpaired t test p = 0.0162. j UMAP plot showing cell clusters from silk-lam VM organoids and k, percentage of cells belonging to each cell cluster from individual organoids at month 4. l Violin plot of percentage of cells belonging to DA neuron cluster from conventional and silk-lam VM organoids at month 4 from three individual organoids. m Expression of selected markers belonging to DA neuron cluster in conventional and silk-lam VM organoids at month 4. Data represent mean ± SEM of 3 biologically independent organoids, two-tailed Wilcoxon Rank Sum test, TH p = 0.0045, SLC6A3 p = 0.010462, KCNQ2 p = 0.0007, ALDH1A1 p = 0.046. Source data are provided as a Source Data file.