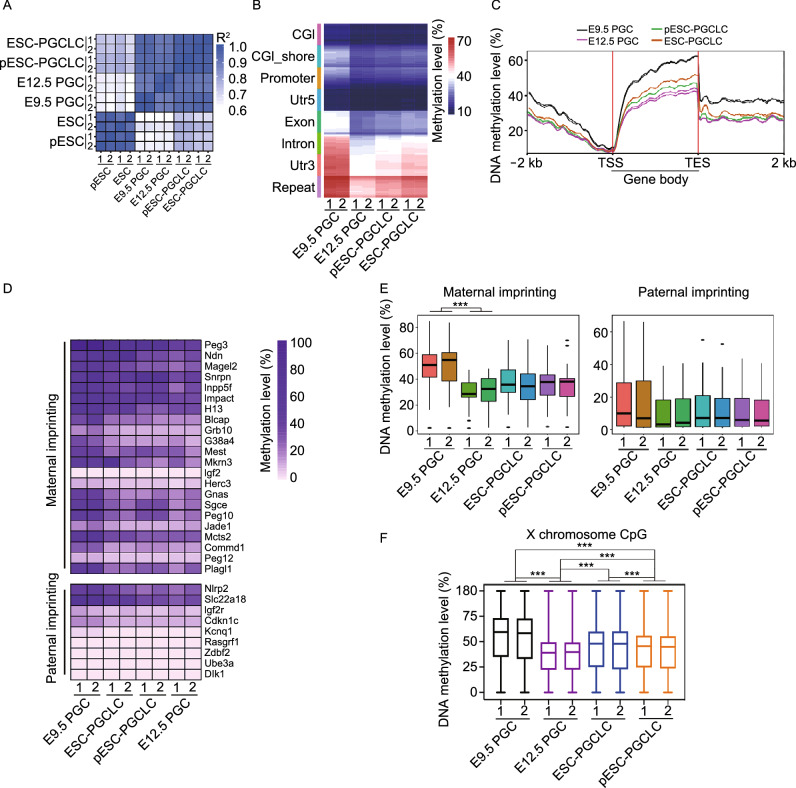
Figure 4.
Comparison of pESC-PGCLCs and PGCs at DNA methylation levels. (A) Correlation factors of genome-wide DNA methylation levels in E9.5 PGCs, E12.5 PGCs, ESC-PGCLCs and pESC-PGCLCs as well as ESCs and pESCs. (B) Heatmap showing genome-wide DNA methylation levels at different gene regions of E9.5 PGCs, E12.5 PGCs, ESC-PGCLCs and pESC-PGCLCs. (C) Genome-wide DNA methylation level in gene body regions including up- and down-stream 2 kb of gene body. (D) Methylation level of maternal and paternal imprinting genes in pESC-PGCLCs, ESC-PGCLCs, E9.5 PGCs and E12.5 PGCs. Methylation counts are provided in Table S1. (E) Box plot showing DNA methylation levels of maternal and paternal imprinting in pESC-PGCLCs, ESC-PGCLCs, E9.5 PGCs and E12.5 PGCs. (F) Box plot showing global DNA methylation levels of X chromosomes in pESC-PGCLCs, ESC-PGCLCs, E9.5 PGCs and E12.5 PGCs. Data shown as mean ± SEM. *P < 0.05, **P < 0.01; ***P < 0.001. RRBS analysis represents two biological replicates