Abstract
In the bio-based polymer industry, putrescine is in the spotlight for use as a material. We constructed strains of Escherichia coli to assess its putrescine production capabilities through the arginine decarboxylase pathway in batch fermentation. N-Acetylglutamate (ArgA) synthase is subjected to feedback inhibition by arginine. Therefore, the 19th amino acid residue, Tyr, of argA was substituted with Cys to desensitize the feedback inhibition of arginine, resulting in improved putrescine production. The inefficient initiation codon GTG of argA was substituted with the effective ATG codon, but its replacement did not affect putrescine production. The essential genes for the putrescine production pathway, speA and speB, were cloned into the same plasmid with argAATG Y19C to form an operon. These genes were introduced under different promoters; lacIp, lacIqp, lacIq1p, and T5p. Among these, the T5 promoter demonstrated the best putrescine production. In addition, disruption of the puuA gene encoding enzyme of the first step of putrescine degradation pathway increased the putrescine production. Of note, putrescine production was not affected by the disruption of patA, which encodes putrescine aminotransferase, the initial enzyme of another putrescine utilization pathway. We also report that the strain KT160, which has a genomic mutation of YifEQ100TAG, had the greatest putrescine production. At 48 h of batch fermentation, strain KT160 grown in terrific broth with 0.01 mM IPTG produced 19.8 mM of putrescine.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13568-021-01330-5.
Keywords: Glutamate-putrescine ligase, N-acetylglutamate synthase, Macrodomain Ori protein, Terrific broth
Introduction
Putrescine is a polyamine and consists of two amino groups and four methylene groups. This compound is widely distributed in living organisms (Pegg 1986; Michael 2016; Keller et al. 2019). Several studies have focused on putrescine as it regulates rapid cell proliferation and differentiation at the level of gene expression (Heby 1981; Michael 2018; Igarashi and Kashiwagi 2000, 2018, 2019). Yoshida et al. (2004) reported a correlation between putrescine and more than 600 genes related to the regulation of transcription. In addition, putrescine is used as a precursor of other polyamines, surfactants, and agrochemicals, and as a component of polymers such as nylon 4,6 (Schneider and Wendisch 2011; Wendisch et al. 2018; Hui et al. 2020). The polycondensation of putrescine and adipic acid is used to synthesize nylon 4,6. Due to its flexibility and high solvent resistance properties, nylon 4,6 was introduced into the commercial field (Demco et al. 2007; Yamanobe et al. 2007). Polyamides are used in the electric vehicle industry as materials to develop a lighter body, interior, motor, controller, and electronic board of the car. Along with the expansion of the global electric vehicle market, the demand for putrescine is also increasing (Scott et al. 2007; Schneider and Wendisch 2010). There are two processes to synthesize putrescine, chemical and biological. However, the chemical process producing an intermediate, succinonitrile, releases dangerous and harmful compounds. On the other hand, the biological process is safe, environmentally friendly, and uses renewable feedstock (Sanders et al. 2007; Nguyen and Lee 2019; Li et al. 2020a).
Among the living organisms that can produce putrescine, bacteria are commonly used in research because they grow rapidly under simple culture conditions such as Escherichia coli. The knowledge of polyamines in E. coli was reviewed admirably by Tabor and Tabor (1985) a long time ago, but it is still valid today. E. coli has two putrescine synthetic pathways (Fig. 1): the ornithine decarboxylase (ODC) pathway and the arginine decarboxylase (ADC) pathway, which use ornithine and arginine as starting compounds, respectively (Fig. 1). Arginine is synthesized from glutamate via ornithine through a sequential reaction. The ODC pathway decarboxylates ornithine to putrescine by constitutive ornithine decarboxylase (SpeC) and/or inducible ornithine decarboxylase (SpeF) (Tabor and Tabor 1985). The other pathway, the ADC pathway, converts arginine to putrescine via agmatine. The conversion of arginine to agmatine is catalyzed by biosynthetic arginine decarboxylase (SpeA) and biodegradative arginine decarboxylase (AdiA). Subsequently, agmatine is hydrolyzed to putrescine by agmatinase (SpeB). Putrescine is catabolized by two pathways, the putrescine utilization pathway (Puu pathway) (Kurihara et al. 2005) and the putrescine aminotransferase (PatA)-γ-aminobutyraldehyde dehydrogenase (PatD) pathway. These catabolic pathways rely on glutamate-putrescine ligase (PuuA) and putrescine aminotransferase (PatA), respectively (Samsonova et al. 2003, 2005; Kurihara et al. 2008). S-Adenosylmethionine decarboxylase (SpeD) generates decarboxylated S-adenosyl-L-methionine. Its n-propylamine is transferred to putrescine via spermidine synthase (SpeE) to synthesize spermidine (Tabor et al. 1986). Spermidine N-acetyltransferase (SpeG) converts spermidine to its inactive form, N-acetylspermidine (Limsuwun and Jones, 2000). In addition, Haywood and Large (1985) reported that speG encodes a putative diamine acetyltransferase that converts putrescine to acetylputrescine in Candida boidinii. Although the use of putrescine by E. coli SpeG as a substrate has not been reported, the speG gene was knocked out in this study to prevent the possible loss of putrescine (Fig. 1). Because of the cationic function of polyamine, it cannot penetrate through the cell membrane. Two spermidine transporters, PotABCD and MdtJI, and six putrescine transporters, PuuP, YdcSTUV, PlaP, PotE, PotFGHI, and SapBCDF, have been identified (Kashiwagi et al. 1992; Pistocchi et al. 1993; Kurihara et al. 2009a, b; Kurihara et al. 2011; Saier et al. 2016; Sugiyama et al. 2016). We recently reported that PotFGHI can import spermidine under biofilm-forming conditions (Thongbhubate et al. 2021).
Fig. 1.
The metabolic map of putrescine in E. coli K-12. The Xs indicate the knocked out genes. Thick arrows indicate the overexpression of genes. Dash arrows indicate the increased expression of genes. Blunt end arrows indicate inhibition of the expression of genes. GdhA glutamate dehydrogenase, HdfR DNA-binding transcriptional dual regulator HdfR, GltBD glutamate synthase, GlnA glutamine synthetase, GlnE glutamine synthetase adenylyltransferase, CarAB carbamoyl phosphate synthetase, ArgA N-acetylglutamate synthase, ArgB acetylglutamate kinase, ArgC N-acetylglutamylphosphate reductase, ArgD N-acetylornithine aminotransferase, ArgE acetylornithine deacetylase, ArgF ornithine carbamoyltransferase, ArgI ornithine carbamoyltransferase, ArgG argininosuccinate synthetase, ArgH argininosuccinate lyase, ArgR DNA-binding transcriptional dual regulator ArgR, SpeA biosynthetic arginine decarboxylase, SpeB agmatinase, SpeC constitutive ornithine decarboxylase, SpeD S-adenosylmethionine decarboxylase, SpeE spermidine synthase, SpeF inducible ornithine decarboxylase, SpeG spermidine N-acetyltransferase, PatA putrescine aminotransferase, PatD γ-aminobutyraldehyde dehydrogenase, GabT 4-aminobutyrate aminotransferase, GabD succinate-semialdehyde dehydrogenase GabD, Sad succinate-semialdehyde dehydrogenase Sad, PuuA glutamate-putrescine ligase, PuuB γ-glutamylputrescine oxidase, PuuC γ-glutamyl-γ-aminobutyraldehyde dehydrogenase, PuuD γ-glutamyl-γ-aminobutyrate hydrolase, PuuP putrescine importer, YdcSTUV putrescine importer, PotFGHI putrescine importer, PlaP putrescine importer, PotE putrescine-ornithine antiporter/putrescine importer, SapBCDF putrescine exporter, TCA cycle tricarboxylic acid cycle
Qian et al. (2009) overproduced putrescine through the ODC pathway in E. coli K-12, and the batch culture experiment resulted in 1.68 g/L (19.0 mM) in the 10 g/L of the glucose-enriched medium. Recently, Li et al. (2020b) overexpressed the ADC pathway and improved putrescine production using E. coli BL21(DE3). They produced putrescine using an SOB medium supplemented with 8 g/L of glucose and 12 g/L of arginine, resulting in 4.77 g/L of putrescine in batch culture. However, the latter method is a type of sequential enzymatic conversion of arginine to putrescine.
We found that the introduction of the plasmid that contains speAB produced more extracellular putrescine than that of the plasmids that contain speC or speABC. Therefore, we focused on fermentation of putrescine via the ADC pathway of E. coli K-12 without the addition of arginine. Here, genetic engineering was used as a tool to increase the production of putrescine. ArgA catalyzes the initial step of arginine biosynthesis (Rajagopal et al. 1998; Shin and Lee 2014; Ginesy et al. 2015). When arginine is overproduced, this compound affects N-acetylglutamate synthase (ArgA) activity through negative feedback inhibition. Furthermore, the DNA-binding transcriptional dual regulator (ArgR) represses the transcription of the genes of arg regulons, including argA. We not only attempted to enhance arginine synthesis, but also deleted genes related to the degradation or utilization of putrescine, in addition to transporters that uptake putrescine from the medium, such as potE, speG, patA, speD, argR and puuPA, and altered the expression of genes related to the synthesis of putrescine, speA, speB, and other genes from plasmids (Fig. 1).
Materials and methods
Construction of E. coli strains
The E. coli strains used in this study are listed in Table 1. The chromosomal genes speD, and argR were replaced with FRT (FLP recombination target)-kanR-FRT from the Keio gene knockout collection (Baba et al. 2006) by P1vir transduction (Miller 1972). The disruption of speG, potE, and the ATP binding site of the puuA gene were described previously (Kurihara et al. 2008, 2009b; Thongbhubate et al. 2021). The patA and potFGHI genes were disrupted by a method described previously (Datsenko and Wanner 2000) using pKD13 as a template for the PCR amplification with delta-ygjG F and delta-ygjG R as primers for patA, and potF-up and potI-down as primers for potFGHI (Additional file 1: Table S1). The kanR genes were eliminated from FRT-kanR-FRT by Flp flippase carried by pCP20 (Datsenko and Wanner 2000). To confirm the deletion of genes, colony PCR was performed with primers (Additional file 1: Table S1) that annealed upstream and downstream of the genomic regions of the target genes, and the sizes of the amplicons were measured. To construct SH2204, the yifE::FRT-kanR-FRT of SH2201 was replaced with the yifEQ100TAG using the zie-296::Tn10 as a co-transduction marker by P1vir phage grown on SH2203 to obtain SH2204, which was confirmed to be KanS. Then, the yifE region of SH2204 was amplified by PCR and the DNA sequence was confirmed. For DNA modification and strain construction, E. coli cells were grown in LB medium (Miller 1972). Where appropriate, culture media were supplemented with 30 μg/mL of kanamycin, 100 μg/mL of ampicillin, or 15 μg/mL of tetracycline. For the pre-cultures, 10 mL of LB medium was inoculated with a single colony in 100-mL Erlenmeyer flasks. Cultures were incubated at 37 °C with reciprocal shaking at 120 rpm.
Table 1.
Strains used in this study
| Strains | Genotype | Source or reference |
|---|---|---|
| FS113 | MG1655 except ΔargR::FRT-kanR-FRT | This study |
| ΔpatA::FRT ΔpotE::FRT ΔspeD::FRT | ||
| ΔspeG::FRT ΔpuuA::FRT | ||
| FS115 | MG1655 except ΔargR::FRT | This study |
| ΔpatA::FRT ΔpotE::FRT ΔspeD::FRT | ||
| ΔspeG::FRT ΔpuuA::FRT | ||
| FS123 | pSH1734/FS113 | This study |
| MG1655 | F− rph-1 | C. A. Gross |
| KN20 | pKN18/FS113 | This study |
| KN24 | pKN11/FS113 | This study |
| KT32 | FS115 except ΔargA::FRT-kanR-FRT | This study |
| KT37 | pSH1823/KT32 | This study |
| KT38 | pSH1807/KT32 | This study |
| KT39 | pSH1820/KT32 | This study |
| KT43 | MG1655 except ΔargR::FRT | This study |
| ΔpatA::FRT ΔpotE::FRT ΔspeD::FRT | ||
| ΔspeG::FRT ΔargA::FRT | ||
| ΔpuuPA::FRT-kanR-FRT | ||
| KT105 | MG1655 except ΔargR::FRT | This study |
| ΔpatA::FRT ΔpotE::FRT ΔspeD::FRT | ||
| ΔspeG::FRT ΔargA::FRT-kanR-FRT | ||
| Strains | Genotype | Source or reference |
| KT112 | MG1655 except ΔargR::FRT | This study |
| ΔpotE::FRT ΔspeD::FRT ΔspeG::FRT | ||
| ΔpuuPA::FRT ΔargA::FRT-kanR-FRT | ||
| KT148 | pKN11/KT32 | This study |
| KT152 | pKN11/KT43 | This study |
| KT153 | pKN11/KT105 | This study |
| KT154 | pKN11/KT112 | This study |
| KT159 | MG1655 except ΔargR::FRT | This study |
| ΔpotE::FRT ΔspeD::FRT ΔspeG::FRT | ||
| ΔargA::FRT-kanR-FRT | ||
| KT160 | pKN11/SH2204 | This study |
| KT161 | pKN11/SH2206 | This study |
| KT162 | pKN11/KT159 | This study |
| KT207 | pSH1820/FS115 | This study |
| KT208 | pKT199/FS115 | This study |
| KT209 | pKT200/FS115 | This study |
| KT210 | pKT201/FS115 | This study |
| SH2201 | MG1655 except ΔargR::FRT | This study |
| ΔpatA::FRT ΔpotE::FRT ΔspeD::FRT | ||
| ΔspeG::FRT ΔargA::FRT | ||
| ΔpuuPA::FRT ΔyifE::FRT-kanR-FRT | ||
| SH2203 | yifEQ100TAG ΔybbA::FRT zie-296::Tn10 This study | |
| SH2204 | MG1655 except ΔargR::FRT | This study |
| ΔpatA::FRT ΔpotE::FRT ΔspeD::FRT | ||
| ΔspeG::FRT ΔargA::FRT | ||
| ΔpuuPA::FRT yifEQ100TAG zie-296::Tn10 | ||
| SH2206 | SH2204 except potFGHI::FRT-kanR-FRT | This study |
Plasmid construction
The plasmids used in this study are shown in Table 2 and the primers used to construct the plasmids are listed in Additional file 1: Table S2. The Wizard Plus SV Minipreps DNA Purification System (Promega; Madison, WI) was used to extract plasmid DNA from cells. PCR amplification was performed using KOD-plus DNA polymerase (Toyobo; Osaka, Japan). The QuikChange method (Stratagene; San Diego, CA) was used to generate argAATG Y19 of pSH1733 using KOD-plus DNA polymerase. There is a 137-base pair space between the speA and speB genes in the genome of MG1655. Therefore, the wild-type speAB genes may not be an operon. The speA and speB genes on the plasmids used in this study are different from the wild-type genes on the genome. They are designed to have only a 21-base pair space with a Shine Dalgarno (SD) sequence between them, i.e., they were designed to be an operon. Consequently, the transcription of speA will also pass through speB. The speC gene was amplified using genomic DNA of MG1655 as a template and primers with EcoRI and SacI recognition sites. After the amplified fragment was cleaved with these two enzymes, it was inserted between the EcoRI and SacI sites of pQE-80L vector.
Table 2.
Plasmids used in this study
| Strains | Genotype | Source or reference |
|---|---|---|
| pCP20 | pSC101 replicon (Ts) bla+ cat+ | Datsenko and Wanner 2000 |
| Flp(γRp) d857 | ||
| pFS29 | pQE-80L except T5p lacO_lacO_speC+ | This study |
| pKD13 | oriRγ bla+ FRT-kanR-FRT | Datsenko and Wanner 2000 |
| pKN11 | pQE-80L except T5p lacO_lacO | This study |
| _speA_speB_argAATG Y19C | ||
| pKN18 | pQE-80L except T5p lacO_lacO | This study |
| _speA_speB_argAATG Y19C T5p lacO_ | ||
| lacO_speC | ||
| pKT199 | pQE-80L except | This study |
| lacIq1p_lacO_speA_speB_argAGTG Y19C | ||
| lacI | ||
| pKT200 | pQE-80L except | This study |
| lacIq1p_lacO_speA_speB_argAATG Y19 | ||
| lacI | ||
| pKT201 | pQE-80L except | This study |
| lacIq1p_lacO_speA_speB_argAGTG Y19 | ||
| lacI | ||
| pQE-80L | ColE1 replicon bla+ lacIq T5p | Qiagen |
| lacO_lacO-(His)6 | ||
| pSH1733 | pQE-80L except argAATG Y19C | This study |
| pSH1734 | pQE-80L except T5p lacO_lacO_speC+ | This study |
| _ argAATG Y19C | ||
| pSH1807 | pQE-80L except | This study |
| lacIqp_lacO_speA_speB_argAATG Y19C | ||
| lacI | ||
| pSH1820 | pQE-80L except | This study |
| lacIq1p_lacO_speA_speB_argAATG Y19C | ||
| lacI | ||
| pSH1823 | pQE-80L except | This study |
| lacIp_lacO_speA_speB_argAATG Y19C | ||
| lacI |
Medium and culture conditions for putrescine production
Pre-culture was carried out in 10 mL of LB medium in a 100-mL Erlenmeyer flask for 16 h at 37 °C with a reciprocal shaking at 120 rpm. After 16 h, the pre-culture was transferred to 10 mL of LB or terrific broth medium to adjust the initial density of the cells to an OD600 of 0.05. The cultures were incubated at 37 °C with a reciprocal shaking at 120 rpm until the turbidity OD600 reached 0.4. Then, IPTG was added to the main culture as needed. When ampicillin was required, 100 μg/mL of ampicillin was added to the media.
Sample preparation
The culture was collected at the indicated time and used to measure the turbidity at OD600. The collected cultured was centrifuged at 18,000 × g at 4 °C for 5 min. The supernatants were collected, and 25 μL of 100% (w/v) trichloroacetic acid was mixed with 250 μL of the three supernatants. Samples were filtrated through a Millex-LH Syringe Driven Filter Unit (Millipore; Billerica, MA).
Measurement of putrescine concentrations
The concentration of putrescine was measured using an LC-20 HPLC device (Shimadzu; Kyoto, Japan) equipped with a TSKgel Polyaminepak (Tosoh; Tokyo, Japan), as described previously (Kurihara et al. 2008). The temperature of the column was set at 50 °C and only one mobile phase solution was used. The flow rate of the mobile phase solution was 0.5 mL/min and that of the detection reagent was 0.42 mL/min. The peak of putrescine was detected at 6 min under this analytical condition. Putrescine was purchased from Nacalai Tesque (Kyoto, Japan). The amounts of polyamines are presented as the average of three independent cultures with standard deviations.
Data analysis
All data were analyzed using SPSS Statistics 25 (IBM; Armonk, NY). The one-way analysis of variance (ANOVA) was used to determine significant differences.
Results
Effects of the initiation codon and Y19C substitution of ArgA
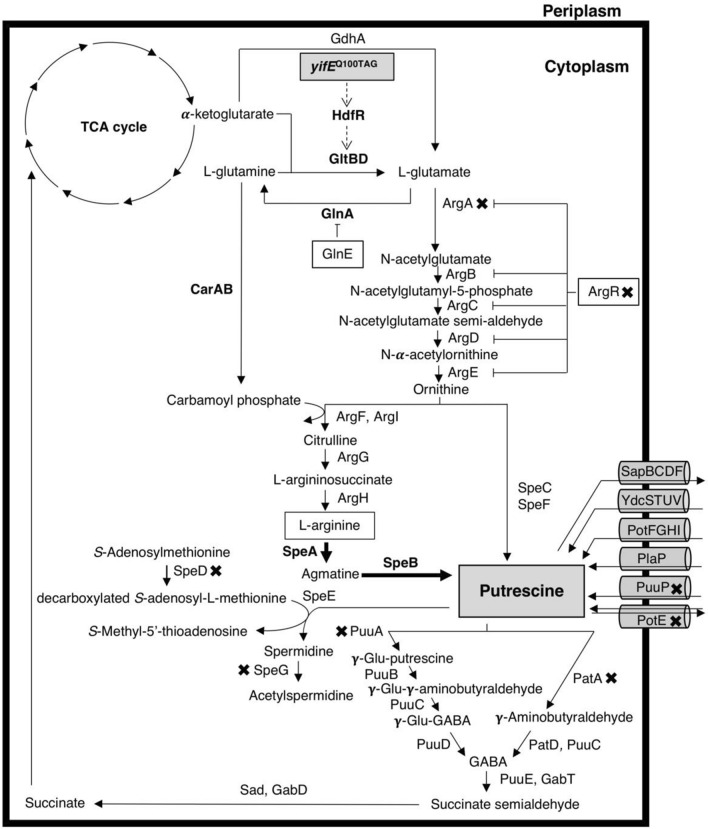
The wild-type strain, MG1655 excretes only 0.18 mM of putrescine in LB medium (data not shown). As arginine is a key substrate of putrescine synthesis through the ADC pathway, to increase putrescine production, we constructed plasmids with the ArgA desensitized to the feedback inhibition by arginine. In addition, the initiation codon of native ArgA is GTG, which is inefficient in translation initiation. Therefore, we generated a plasmid containing the argA gene with the effective ATG codon. Moreover, speA and speB genes were introduced under the lacIq1 promoter along with the argA gene in order to improve putrescine production. To compare the effects of both amino acid and initiation codon substitution, pSH1820 (argAATG Y19C), pKT199 (argAGTG Y19C), pKT200 (argAATG Y19), and pKT201 (argAGTG Y19) were constructed, and strain FS115 was transformed to obtain KT207, KT208, KT209, and KT210, respectively. The strains were grown in LB medium with ampicillin and incubated at 37 °C. The result is illustrated in Fig. 2. KT207 excreted the highest concentration of putrescine into the medium, whereas strain KT210 excreted the lowest amount of putrescine. To compare the effects of the initiation codon, the strains were separated into two groups: GTG codon (KT208 and KT210) and ATG codon (KT207 and KT209). ATG codon exhibited a higher concentration of putrescine than the original initiation codon, GTG. In addition, Y19C substitution in strains KT207 and KT208 resulted in notably higher putrescine production than strains containing the native amino acid residue Y19 (KT209 and KT210).
Fig. 2.
Base substitution on argA increased putrescine production. Putrescine concentration in the supernatant of strains harboring plasmids with different mutant argA: KT207 (argAATG Y19C), KT208 (argAGTG Y19C), KT209 (argAATG Y19), and KT210 (argAGTG Y19). The strains were grown in LB medium supplemented with 100 μg/mL of ampicillin at 37 °C for 48 h with reciprocal shaking at 120 rpm. When the OD600 reached 0.4, 0.02 mM IPTG was added. **, p < 0.01; ***, p < 0.001. Data shown are averages ± standard deviations, and culture experiments were performed in triplicate
Effect of SpeA, SpeB, and SpeC on putrescine production
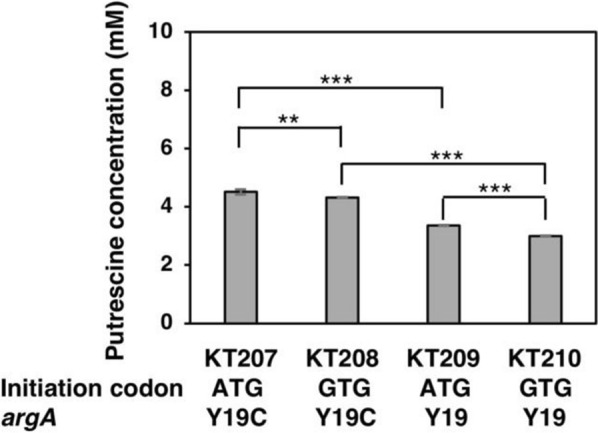
There are ADC and ODC pathways for putrescine synthesis in E. coli. Whether the over-expression of ADC pathway only or of both ADC and ODC pathways is appropriate for the over-production of putrescine was not sure. To address this issue, we constructed a plasmid that has argAATG Y19C together with only speC gene under T5 promoter (pFS29) and compare with another two plasmids which have argAATG Y19C with only speAB genes (pKN11) and argAATG Y19C with speAB and speC genes (pKN18). The plasmids were transformed into MG1655 as a host strain, resulting in FS123, KN24, and KN20, respectively. As depicted in Fig. 3A, the cell growth profiles of the above three strains were not different. However, the putrescine production of KN24, which has only speAB genes on a plasmid with argAATG Y19C under T5 promoter, exhibited the highest putrescine content among these three strains (Fig. 3B). Therefore, the plasmid pKN11, which contains only speAB genes with argAATG Y19C, was selected to use in further experiments.
Fig. 3.
Effect of SpeA, SpeB, and SpeC on putrescine production. A Bacterial cell growth and B putrescine concentration in the supernatant of strains FS123 (speC+, open circle), KN20 (speABC+, closed triangle), and KN24 (speAB+, closed square) grown in LB medium supplemented with 100 μg/mL of ampicillin. When the OD600 reached 0.4, 0.02 mM IPTG was added. Data shown are averages ± standard deviations, and culture experiments were performed in triplicate
Effects of wild-type ArgA on ArgAATG Y19C
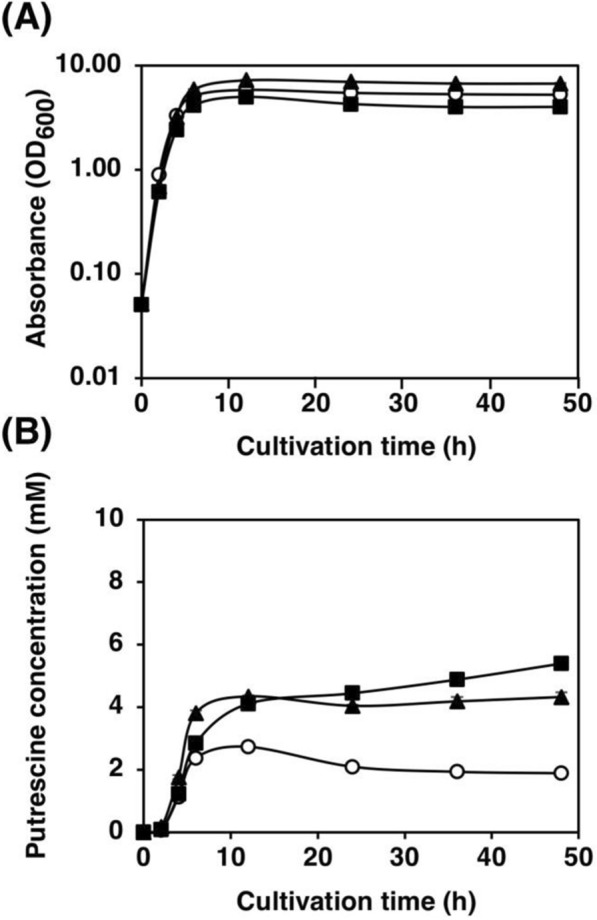
ArgA forms a homohexamer to be an active enzyme. When genomic argA is wild-type, it may form a mixed hexamer of ArgAwt and ArgAY19C. ArgAwt may be dominant to ArgAY19C in the desensitization of arginine. To compare the effects of wild-type ArgA, the strains with and without argA, FS115 (argA+) and KT32 (ΔargA), respectively, were constructed. The plasmid carrying speAB and argA (argAATG Y19C) encoding desensitized ArgAATG Y19C under the regulation of the lacIq1 promoter and lac operator was constructed (pSH1820) and used to transform the above-mentioned strains (FS115 and KT32), resulting in KT207 and KT39, respectively. To evaluate the effects of the coexistence of wild-type ArgA and desensitized ArgAATG Y19C, KT207 and KT39 were used. As depicted in Fig. 4, the growth profile and putrescine production did not significantly differ between KT207 and KT39.
Fig. 4.
Effects of the wild-type ArgA expressed from the genome on putrescine production. A Bacterial cell growth and B putrescine concentration in the supernatant of strains KT39 (ΔargA, open circle) and KT207 (argA+, closed triangle) grown in LB medium supplemented with 100 μg/mL of ampicillin. When the OD600 reached 0.4, 0.02 mM IPTG was added. Data shown are averages ± standard deviations, and culture experiments were performed in triplicate
Effects of promoter strength on putrescine production
To promote the sequential reactions from arginine to putrescine, the promoter for speAB_argAATG Y19C was evaluated. The concentrations of IPTG for inducible lacI, lacIq, lacIq1, and T5 promoters were compared. The lacI promoter of pSH1823 was replaced by lacIq and lacIq1promoters, resulting in pSH1807 and pSH1820, respectively. The plasmid (pKN11) carrying the speAB_argAATG Y19C under the T5 promoter and two lac operators was constructed. Strain KT32 was transformed with each plasmid to obtain KT37 (lacIp), KT38 (lacIqp), KT39 (lacIq1p), and KT148 (T5p). These strains were grown in LB medium with the addition of 0.02 mM IPTG. The putrescine production of KT38 (lacIqp) was not different from that of KT37 (lacIp). On the other hand, strains KT39 (lacIq1p), and KT148 (T5p) produced 349% and 431% more putrescine than strain KT37 (lacIp), respectively (Fig. 5B). Based on these results, KT148, carrying speAB and desensitized argA under the T5 promoter and two lac operators, was used for further study. To optimize the IPTG concentration, it was varied from 0 to 0.20 mM. Among these concentrations, 0.03 mM IPTG resulted in the highest putrescine production by KT148 in LB medium (data not shown).
Fig. 5.
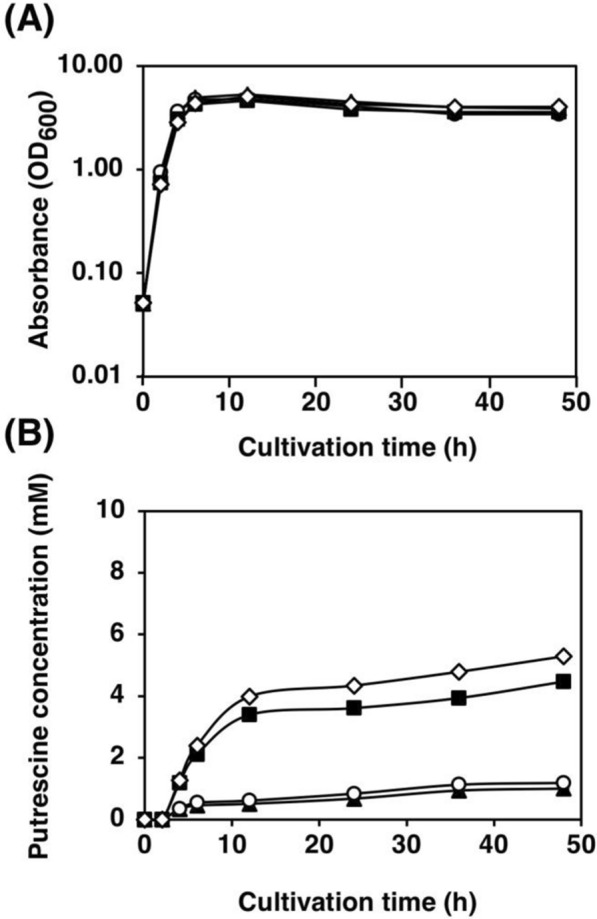
Effects of promoters of speAB and desensitized argA on putrescine production. A Bacterial cell growth and B putrescine concentration in the supernatant of strains KT37 (lacI promoter, closed triangle), KT38 (lacIq promoter, open circle), KT39 (lacIq1 promoter, closed square), and KT148 (T5 promoter, open diamond) grown in LB medium supplemented with 100 μg/mL of ampicillin. When the OD600 reached 0.4, 0.02 mM IPTG was added. Data shown are averages ± standard deviations, and culture experiments were performed in triplicate
Disruption of genomic puuPA increases putrescine production
To improve the accumulation of extracellular putrescine, the importer and degradation enzymes of putrescine should be eliminated. The host strains KT32 (ΔpuuA ΔpatA), KT43 (ΔpuuAP ΔpatA), KT105 (puuP+A+ ΔpatA), KT112 (ΔpuuAP patA+), and KT159 (puuP+A+ patA+) were constructed to assess the effects of patA, puuA, and puuP on putrescine production. The above strains were transformed by the plasmid pKN11 (T5p_speAB_argAATG Y19C), leading to strains KT148, KT152, KT153, KT154, and KT162. Among all strains, puuP+A+ strains (KT153 and KT162) produced a lower amount of putrescine than the ΔpuuA (KT148) and ΔpuuAP (KT152 and KT154) strains (Fig. 6B). However, there were no significant differences in the concentration of extracellular putrescine between KT153 (ΔpatA) and KT162 (patA+) strains.
Fig. 6.
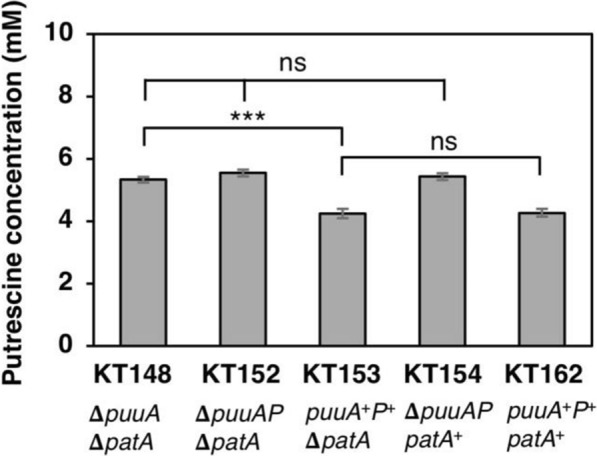
Effects of patA, puuA, and puuP genes on the putrescine production. The amount of putrescine in the culture supernatant of the strains KT148, KT152, KT153, KT154, and KT162 grown in LB medium supplemented with 100 μg/mL of ampicillin for 48 h. When the OD600 reached 0.4, 0.03 mM IPTG was added. ***, p < 0.001; ns, not significantly different. Data shown are averages ± standard deviations, and culture experiments were performed in triplicate
Effects of the medium on putrescine production
The amount of putrescine production was compared using LB medium and terrific broth to optimize the medium for putrescine production. Strain KT152 grew and produced putrescine more in terrific broth than in LB (Fig. 7). At 48 h of incubation, strain KT152 excreted 10.9 mM of extracellular putrescine in terrific broth, which was about twice as much as that in LB (Fig. 7B). Moreover, the IPTG concentration was optimized for cultivation in terrific broth. The concentration of IPTG was varied from 0 to 0.20 mM. The optimal concentration of IPTG for KT152 cultured in terrific broth was 0.02 mM (data not shown).
Fig. 7.
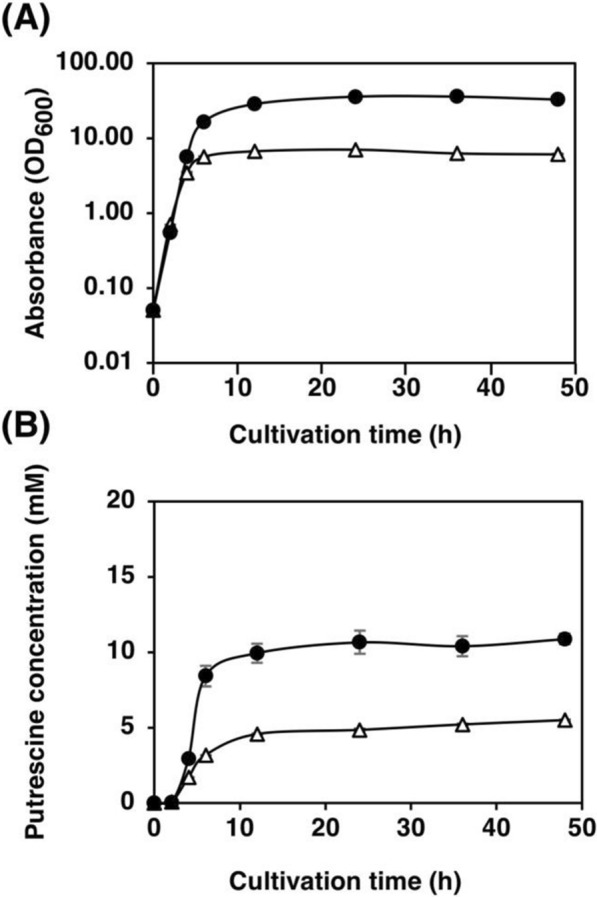
Effects of culture media on putrescine production by strain KT152. A Bacterial cell growth and B putrescine concentration in the supernatant of the strain KT152, whose putrescine utilization pathway (ΔpuuAP ΔpatA) was disrupted. Open triangles represent the strain KT152 grown in the LB medium supplemented with 100 μg/mL of ampicillin; closed circles represent the strain KT152 grown in terrific broth supplemented with 100 μg/mL of ampicillin. When the OD600 reached 0.4, 0.03 mM IPTG was added. Data shown are averages ± standard deviations, and culture experiments were performed in triplicate
Effects of putrescine transporter PotFGHI and mutant YifEQ100TAG
To clarify whether the disruption of the PotFGHI transporter and the mutation on YifE improved putrescine production, strain SH2204 was transformed with yifE::FRT-kanR-FRT of SH2201 and the yifEQ100TAG was replaced using zie-296::Tn10 as a co-transduction marker by P1vir phage grown on SH2203. The potFGHI operon of the strain SH2204 (yifEQ100TAG) was disrupted by P1vir transduction to obtain strain SH2206 (yifEQ100TAG ΔpotFGHI). Strains KT43, SH2204, and SH2206 were transformed with pKN11 (T5p_speAB_argAATG Y19C), and strains KT152, KT160, and KT161 were obtained. Putrescine production by these strains in a terrific broth with the addition of 0.02 mM IPTG is compared in Fig. 8a. Samples were taken at 0, 2,4, 6, 12, 24, 36, and 48 h of incubation. Among these sampling times, the highest concentration of putrescine was observed at 48 h (data not shown). There was no significant difference in putrescine production between strains KT160 and KT161. However, the amount of extracellular putrescine excreted from both KT160 (yifEQ100TAG potFGHI+) and KT161 (yifEQ100TAG ΔpotFGHI) was 23% higher than that by KT152 (yifE+ potFGHI+) (Fig. 8a). In addition, the IPTG concentration was optimized from 0 to 0.2 mM for the cultivation of KT160 in terrific broth. The optimal concentration of IPTG for KT160 cultured in terrific broth was 0.01 mM and the strain produced 19.8 mM of putrescine (Fig. 8b).
Fig. 8.
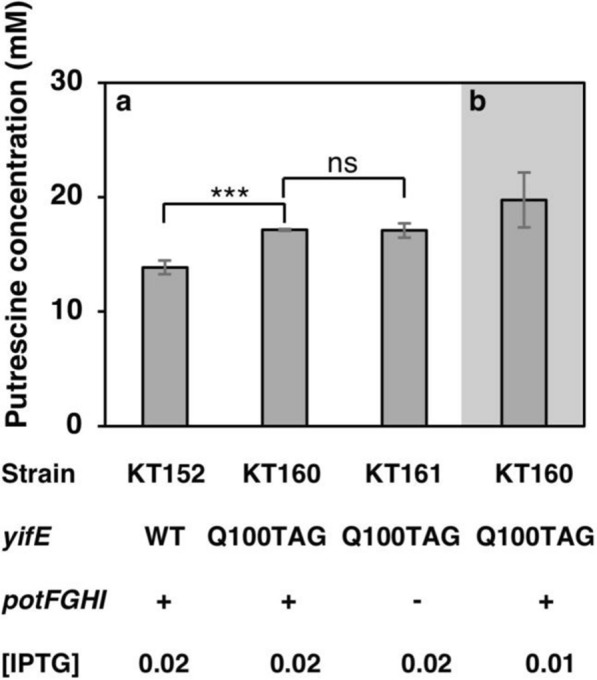
Effects of the yifE mutant and putrescine transporter PotFGHI on putrescine production. a The putrescine concentration in the supernatant of the culture media of strains KT152, KT160, and KT161 grown in terrific broth supplemented with 100 μg/mL of ampicillin for 48 h. When the OD600 reached 0.4, 0.02 mM IPTG was added. b The putrescine concentration in the supernatant of the culture media of strains KT160 grown in terrific broth supplemented with 100 μg/mL of ampicillin. When the OD600 reached 0.4, 0.01 mM IPTG was added. * p < 0.001; ns, not significantly different. yifE: WT, yifEwt; Q100TAG, yifEQ100TAG gene. potFGHI: + , presence of potFGHI operon; -, absence of potFGHI operon. [IPTG]: 0.01 and 0.02 is the optimum concentration of IPTG in terms of mM. Data shown are averages ± standard deviations, and culture experiments were performed in triplicate
Discussion
Efficient initiation codon of argAY19C improved the putrescine production- Arginine is a key substrate of putrescine synthesis through the ADC pathway. The biosynthetic pathway of arginine initiates with glutamate to N-acetylglutamate by N-acetylglutamate synthase (ArgA), which is encoded by argA. However, not only is the transcription of the argA gene repressed by ArgR, but the activity of ArgA is also inhibited by the final product arginine (Lu 2006; Sun et al. 2015; Xu and Zhang 2019a, b). To produce putrescine from glutamate, the repression of argA by ArgR and the feedback inhibition of ArgA by arginine are of concern. Previously, Rajagopal et al. (1998) constructed plasmids with a point mutation in the argA gene. Among all point mutations, the strain with the Y19C mutation, which is an amino acid substitution of tyrosine with cysteine at the 19th codon, was able to produce the highest amount of arginine. According to the review of Igarashi and Kashiwagi (2010), the addition of putrescine stimulates the translation of polyamine modulons. As ArgA is a polyamine modulon (Igarashi et al. 2015), its translation can be stimulated by the increase in putrescine. The effects of the substitution of the GTG codon with the ATG codon was evaluated, but the substitution of GTG with ATG was not so effective as expected (Fig. 2). This may be because the plasmid we constructed has only 8 base pairs between the SD sequence and the ATG initiation codon, and it lacks the characteristics of a polyamine modulon. However, desensitization of ArgA by amino acid substitution at 19 positions from tyrosine to cysteine from the plasmid increased putrescine production (Fig. 2). And the strain with argAATG Y19C had the highest amount of putrescine.
The effects of wild-type ArgA on desensitized ArgA- To confirm whether ArgAwt or ArgAATG Y19C is dominant, we compared the putrescine production of KT39 (argAATG Y19C/ΔargA) and KT207 (argAATG Y19C/argA+) strains. The growth and putrescine production of both KT39 and KT207 were not different. Thus, the presence of wild-type ArgA expressed from the genome does not affect the growth or putrescine production (Fig. 4).
The strong promoter T5p increases putrescine production- According to the report of Buch and Boyle (1985), a significant proportion of SpeA associates with the cell envelope. A recent paper reported that some of SpeA is in the periplasm and the other is in the cytoplasm (Meydan et al. 2019). Overexpression of membrane-associated or excreted proteins can affect cell health. Therefore, in this study, the promoter strength required for overexpression was of concern. The lacI, lacIq, lacIq1, and T5 promoters were compared. The strain carrying the lacI promoter produced the lowest concentration of extracellular putrescine, whereas the strain carrying the T5 promoter produced the highest. Due to its affinity for RNA polymerase, the lacI promoter is the poorest (Muller-Hill et al. 1968). Therefore, the expression level is low. A point mutation at the − 35 region of the lacIq promoter led to tenfold higher expression and the lacIq1 promoter resulted in 170-fold higher expression (Calos 1978; Glascock and Weickert 1998; Penumetcha et al. 2010). Moreover, there are two lac operators in this T5 promoter with a strong affinity to E. coli RNA polymerase (von Gabain and Bujard 1977, 1979; Shibui et al. 1988), but no study has directly compared their affinity for RNA polymerase. As depicted in Fig. 5B, the putrescine production of KT148 (T5p) was higher than that of KT39 (lacIq1p).
Effects of puuPA disruption on putrescine production- Two catabolic pathways of putrescine have been reported. PatA and PuuA are the first enzymes of the aminotransferase pathway and putrescine utilization pathway (the Puu pathway), respectively. PatA transfers one of the amino groups of putrescine to α-ketoglutarate (Shaibe et al. 1985; Schneider and Reitzer 2012). Another enzyme that catabolizes putrescine is PuuA, which γ-glutamylates the amino group of putrescine to generate γ-glutamylputrescine. The proteins involved in the Puu pathway are encoded in a gene cluster. The importer of this Puu pathway is PuuP, which is encoded in this gene cluster. After PuuP transports putrescine into the cell, the degradation step begins with PuuA (Kurihara et al. 2009b). There is no report to compare the reaction velocity or the expression level between the puuA and patA yet. So, it is difficult to specify which enzymes are mainly work. However, according to our previous study, the deletion of the puuA gene caused a severe effect on putrescine utilization and ΔpuuA strain could not grow using putrescine as a nitrogen source or a carbon source, while the deletion of the patA gene did not affect this issue (Kurihara et al. 2008). The result in this study is agreeable with the previous study that the presence of puuA showed the degradation of putrescine while patA is not. In addition, under the condition in which putrescine is present, PuuP is required as a putrescine transporter to use putrescine as an energy source (Kurihara et al. 2009b; Terui et al. 2014). As PuuP may also have its own promoter, we constructed KT148 (∆puuA) and KT152 (∆puuAP) to investigate the relationship between these puuAP genes. As shown in Fig. 6, puuAP may be an operon because ∆puuAP had the same result as ∆puuA. The deletion of puuA increased the extracellular accumulation of putrescine. In summary, PatA does not affect putrescine production, but the deletion of PuuA increases the amount of putrescine.
Terrific broth is a better medium for putrescine production- LB medium is widely used as a standard medium for a broad group of bacteria (Sezonov et al. 2007; Suzuki et al., 2019). However, the growth of E. coli is not high in the LB medium, whereas it is markedly high in terrific broth (Losen et al. 2004; Lessard 2013). Terrific broth contains 5.04 g/L of glycerol, while LB medium does not. E. coli can utilize glycerol as a carbon source for growth and putrescine production. Besides much higher concentration of yeast extracts, this is one of the reasons why the growth and putrescine production in terrific broth is better than those in LB medium (Fig. 7).
Mutation of YifE increases putrescine- During we attempted to find a putrescine exporter in E. coli K-12, we found that the extracellular putrescine concentration of strain JW0484 with ΔybbA::FRT-kanR-FRT was approximately double that of its parental strain, ME9062 (Additional file 1: Fig. S1). The ΔybbA::FRT-kanR-FRT was transduced to strain MG1655 to confirm the increase of putrescine production. However, the extracellular putrescine concentration of the MG1655 with ΔybbA::FRT-kanR-FRT was not different from that of MG1655. We predicted that the second mutation in strain JW0484 increased the extracellular putrescine concentration. Then, its genome sequence was performed to evaluate this prediction. The result of genome sequencing showed several mutations in strain JW0484, which were not reported in strain ME9062. Additional mutations were found in intA, fbaA, dgoR, yifE, and lexA in the genome of strain JW0484. Further experiments revealed that mutation of the yifE gene increases the extracellular putrescine concentration (Additional file 1: Fig. S2). As shown in Fig. 8a, the YifE mutant was also effective in our putrescine over-producing strain (KT160). RT-PCR revealed that the yifEQ100TAG mutation increases the transcription of hdfR and gltB genes.
Pistocchi et al. (1993) reported that the PotFGHI transporter uptakes putrescine and accumulates it in the cell. Therefore, it is conceivable that PotFGHI reduces extracellular putrescine. We compared the strains with and without the PotFGHI transporter, but there was no difference in the concentration of putrescine between potFGHI+ and ΔpotFGHI strains. Thus, the PotFGHI transporter does not affect the amount of extracellular putrescine. Terui et al. (2014) reported that PotFGHI functions as a major transporter when putrescine is absent in the medium. However, the PotFGHI transporter can be inhibited by putrescine through a feedback loop. In our experimental condition, putrescine was synthesized and exported to the medium; therefore, PotFGHI may have been inhibited by putrescine. We achieved 19.8 mM of putrescine production from KT160 in terrific broth. When compare in terms of the product yield, Qian et al. (2009) reported a yield of putrescine at 30.3 g/M glucose, while our putrescine production is the 31.8 g/M glycerol. However, as putrescine is an ionic compound, its production altered the pH of the medium during cultivation. As we do not have a bioreactor device, we were unable to assess continuous feeding with pH adjustment.
Supplementary Information
Additional file 1: Table S1. Primers used for strain construction. Table S2. Primers used for plasmid construction. Table S3. Primers used for Realtime PCR. Fig. S1. Comparison of extracellular putrescine concentration per OD600 among strains of the Keio collection. M9 glucose medium (60 mL in 100-mL Erlenmeyer flask with stirrer bar) was inoculated with the pre-culture at the initial OD600 of 0.06. The flasks were set in the 7-L rectangular anaerobic jar with 2 AnaeroPack-Anaero sachets (Mitsubishi Gas Chemical; Tokyo, Japan) to form an anaerobic environment and the jar was set on the stirrer in the microbiological incubator. The culture medium was stirred at 150 rpm and kept at 37°C. After 10 h of incubation, the OD600 of the culture was measured and 0.4 mL of the culture was centrifuged. Thirty μL of 100% (w/v) TCA was mixed with 0.3 mL of the supernatant, filtrated through the membrane filter, and then subjected to HPLC analysis as described in the main text. Asterisks indicate the genes with significant differences in transcription levels. Fig. S2. Q100TAG mutation in the yifE gene promotes putrescine production. Extracellular concentration of putrescine of AI32 (pQE-80L/MG1655, open diamond) and AI33 (pQE-80L::yifEQ100TAG/MG1655, closed square) cultured at 37°C in minimal M9 supplemented with 0.2% glucose. 100 μg/mL of ampicillin was added to maintain the plasmids. When the OD600 reached 0.5, the 0.5 mM IPTG was added. Fig. S3. The presence of hdfR had a slight, if any, effect on the increase in extracellular putrescine concentration by YifEQ100TAG. (A) Cell growth and (B) extracellular concentration of putrescine of AI32 (pQE-80L/MG1655, open circle), AI33 (pQE-80L::yifEQ100TAG/MG1655, closed triangle), and KT218 (pQE-80L::yifEQ100TAG/ MG1655 but ΔhdfR::FRT-kanR-FRT, closed square) cultured at 37°C in LB supplemented with 100 μg/mL of ampicillin. When the OD600 reached 0.4, 0.02 mM IPTG was added.
Acknowledgements
Not applicable.
Authors’ contributions
KT conducted all fermentation experiments in the main text and contributed to the interpretation, analyzation and preparation of the draft. KI constructed a plasmid and performed the primary fermentation experiments. YS and AI identified the yifE mutant and found that the mutation can increase putrescine production. HS planned the study, acquired the funding, led the project, constructed some strains and plasmids, and edited the manuscript.
Funding
This study was partially supported by funding from the Kinugasa Research Foundation for Textile Science.
Availability of data and materials
Some data are not shown in the text and supplement. However, they are available from the authors upon reasonable request. The strains and plasmids used in this work can be shared by exchanging MTA.
Declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing of interests
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch JK, Boyle SM. Biosynthetic arginine decarboxylase in Escherichia coli is synthesized as a precursor and located in the cell envelope. J Bacteriol. 1985;163:522–527. doi: 10.1128/jb.163.2.522-527.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos MP. DNA sequence for a low-level promoter of the lac repressor gene and an ‘up’ promoter mutation. Nature. 1978;274:762–765. doi: 10.1038/274762a0. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demco DE, Litvinov VM, Rata G, Popescu C, Phan KH, Schmidt A, Blümich B. Investigation of thermal aging of polyamide 4, 6 by 1H solid-state NMR. Macromol Chem Phys. 2007;208:2085–2095. doi: 10.1002/macp.200700095. [DOI] [Google Scholar]
- Ginesy M, Belotserkovsky J, Enman J, Isaksson L, Rova U. Metabolic engineering of Escherichia coli for enhanced arginine biosynthesis. Microb Cell Factories. 2015;14:29. doi: 10.1186/s12934-015-0211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glascock CB, Weickert MJ. Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli. Gene. 1998;223:221–231. doi: 10.1016/s0378-1119(98)00240-6. [DOI] [PubMed] [Google Scholar]
- Haywood GW, Large PJ. The occurrence, subcellular localization and partial purification of diamine acetyltransferase in the yeast Candida boidinii grown on spermidine or putrescine as sole nitrogen source. Eur J Biochem. 1985;148:277–283. doi: 10.1111/j.1432-1033.1985.tb08836.x. [DOI] [PubMed] [Google Scholar]
- Heby O. Role of polyamines in the control of cell proliferation and differentiation. Differentiation. 1981;19:1–20. doi: 10.1111/j.1432-0436.1981.tb01123.x. [DOI] [PubMed] [Google Scholar]
- Hui H, Bai Y, Fan TP, Zheng X, Cai Y. Biosynthesis of putrescine from L-arginine using engineered Escherichia coli whole cells. Catalysts. 2020;10:947. doi: 10.3390/catal10090947. [DOI] [Google Scholar]
- Igarashi K, Terui Y, Kashiwagi K. The polyamine modulon: genes encoding proteins whose synthesis is enhanced by polyamines at the level of translation. In: Kusano T, Suzuki H, editors. Polyamines: a universal molecular nexus for growth, survival, and specialized metabolism. Tokyo: Springer; 2015. pp. 131–1411. [Google Scholar]
- Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Modulation of cellular function by polyamines. Int J Biochem Cell Biol. 2010;42:39–51. doi: 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Effects of polyamines on protein synthesis and growth of Escherichia coli. J Biol Chem. 2018;293:18702–18709. doi: 10.1074/jbc.TM118.003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. The functional role of polyamines in eukaryotic cells. Int J Biochem Cell Biol. 2019;107:104–115. doi: 10.1016/j.biocel.2018.12.012. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:4529–4533. doi: 10.1073/pnas.89.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Chattopadhyay M, Tabor H. Absolute requirement for polyamines for growth of Escherichia coli mutants (mnmE/G) defective in modification of the wobble anticodon of transfer-RNA. FEMS Microbiol Lett. 2019;366:fnz110. doi: 10.1093/femsle/fnz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara S, Oda S, Kato K, Kim HG, Koyanagi T, Kumagai H, Suzuki H. A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J Biol Chem. 2005;280:4602–4608. doi: 10.1074/jbc.M411114200. [DOI] [PubMed] [Google Scholar]
- Kurihara S, Oda S, Tsuboi Y, Kim HG, Oshida M, Kumagai H, Suzuki H. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J Biol Chem. 2008;283:19981–19990. doi: 10.1074/jbc.M800133200. [DOI] [PubMed] [Google Scholar]
- Kurihara S, Suzuki H, Tsuboi Y, Benno Y. Dependence of swarming in Escherichia coli K-12 on spermidine and the spermidine importer. FEMS Microbiol Lett. 2009;294:97–101. doi: 10.1111/j.1574-6968.2009.01552.x. [DOI] [PubMed] [Google Scholar]
- Kurihara S, Tsuboi Y, Oda S, Kim HG, Kumagai H, Suzuki H. The putrescine importer PuuP of Escherichia coli K-12. J Bacteriol. 2009;191:2776–2782. doi: 10.1128/JB.01314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara S, Suzuki H, Oshida M, Benno Y. A novel putrescine importer required for type 1 pili-driven surface motility induced by extracellular putrescine in Escherichia coli K-12. J Biol Chem. 2011;286:10185–10192. doi: 10.1074/jbc.M110.176032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JC. Growth media for E. coli. Methods Enzymol. 2013;533:181–189. doi: 10.1016/B978-0-12-420067-8.00011-8. [DOI] [PubMed] [Google Scholar]
- Li L, Zou D, Ji A, He Y, Liu Y, Deng Y, Chen S, Wei X. Multilevel metabolic engineering of Bacillus amyloliquefaciens for production of the platform chemical putrescine from sustainable biomass hydrolysates. ACS Sustain Chem Eng. 2020;8:2147–2157. doi: 10.1021/acssuschemeng.9b05484. [DOI] [Google Scholar]
- Li G, Huang D, Wang L, Deng Y. Highly efficient whole-cell biosynthesis of putrescine by recombinant Escherichia coli. Biochem Eng J. 2020;166:107859. doi: 10.1016/j.bej.2020.107859. [DOI] [Google Scholar]
- Limsuwun K, Jones PG. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J Bacteriol. 2000;182:5373–5380. doi: 10.1128/JB.182.19.5373-5380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losen M, Frölich B, Pohl M, Büchs J. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol Prog. 2004;20:1062–1068. doi: 10.1021/bp034282t. [DOI] [PubMed] [Google Scholar]
- Lu CD. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl Microbiol Biotechnol. 2006;70:261–272. doi: 10.1007/s00253-005-0308-z. [DOI] [PubMed] [Google Scholar]
- Meydan S, Marks J, Klepacki D, Sharma V, Baranov PV, Firth AE, Margus T, Kefi A, Vázquez-Laslop N, Mankin AS. Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol Cell. 2019;74:481–493. doi: 10.1016/j.molcel.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AJ. Polyamines in eukaryotes, bacteria, and archaea. J Biol Chem. 2016;291:14896–14903. doi: 10.1074/jbc.R116.734780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AJ. Polyamine function in archaea and bacteria. J Biol Chem. 2018;293:18693–18701. doi: 10.1074/jbc.TM118.005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Müller-Hill B, Crapo L, Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968;59:1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Lee EY. Biological conversion of methane to putrescine using genome-scale model-guided metabolic engineering of a methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. Biotechnol Biofuels. 2019;12:147. doi: 10.1186/s13068-019-1490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penumetcha P, Lau K, Zhu X, Davis K, Eckdahl TT, Campbell AM. Improving the Lac system for synthetic biology. Bios. 2010;81:7–15. doi: 10.1893/011.081.0104. [DOI] [Google Scholar]
- Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem. 1993;268:146–152. doi: 10.1016/S0021-9258(18)54126-0. [DOI] [PubMed] [Google Scholar]
- Qian ZG, Xia XX, Lee SY. Metabolic engineering of Escherichia coli for the production of putrescine: a four carbon diamine. Biotechnol Bioeng. 2009;104:651–662. doi: 10.1002/bit.22502. [DOI] [PubMed] [Google Scholar]
- Rajagopal BS, DePonte J, Tuchman M, Malamy MH. Use of inducible feedback-resistant N-acetylglutamate synthetase (argA) genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains. Appl Environ Microbiol. 1998;64:1805–1811. doi: 10.1128/AEM.64.5.1805-1811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 2016;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonova NN, Smirnov SV, Altman IB, Ptitsyn LR. Molecular cloning and characterization of Escherichia coli K12 ygjG gene. BMC Microbiol. 2003;3:2. doi: 10.1186/1471-2180-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonova NN, Smirnov SV, Novikova AE, Ptitsyn LR. Identification of Escherichia coli K12 YdcW protein as a γ-aminobutyraldehyde dehydrogenase. FEBS Lett. 2005;579:4107–4112. doi: 10.1016/j.febslet.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Sanders J, Scott E, Weusthuis R, Mooibroek H. Bio-refinery as the bio-inspired process to bulk chemicals. Macromol Biosci. 2007;7:105–117. doi: 10.1002/mabi.200600223. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Reitzer L. Pathway and enzyme redundancy in putrescine catabolism in Escherichia coli. J Bacteriol. 2012;194:4080–4088. doi: 10.1128/JB.05063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Wendisch VF. Putrescine production by engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2010;88:859–868. doi: 10.1007/s00253-010-2778-x. [DOI] [PubMed] [Google Scholar]
- Schneider J, Wendisch VF. Biotechnological production of polyamines by bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol. 2011;91:17–30. doi: 10.1007/s00253-011-3252-0. [DOI] [PubMed] [Google Scholar]
- Scott E, Peter F, Sanders J. Biomass in the manufacture of industrial products-the use of proteins and amino acids. Appl Microbiol Biotechnol. 2007;75:751–762. doi: 10.1007/s00253-007-0932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezonov G, Joseleau-Petit D, D'Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaibe E, Metzer E, Halpern YS. Metabolic pathway for the utilization of L-arginine, L-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J Bacteriol. 1985;163:933–937. doi: 10.1128/jb.163.3.933-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibui T, Uchida M, Teranishi Y. A new hybrid promoter and its expression vector in Escherichia coli. Agr Biol Chem. 1988;52:983–988. doi: 10.1080/00021369.1988.10868770. [DOI] [Google Scholar]
- Shin JH, Lee SY. Metabolic engineering of microorganisms for the production of L-arginine and its derivatives. Microb Cell Factories. 2014;13:166. doi: 10.1186/s12934-014-0166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Nakamura A, Matsumoto M, Kanbe A, Sakanaka M, Higashi K, Igarashi K, Katayama T, Suzuki H, Kurihara S. A novel putrescine exporter SapBCDF of Escherichia coli. J Biol Chem. 2016;291:26343–26351. doi: 10.1074/jbc.M116.762450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Song W, Liu L. Enzymatic production of agmatine by recombinant arginine decarboxylase. J Mol Catal B Enzym. 2015;121:1–8. doi: 10.1016/j.molcatb.2015.06.008. [DOI] [Google Scholar]
- Suzuki H, Nishida K, Tamaki H. Shochu slop is an excellent medium for Escherichia coli K-12. Lett Appl Microbiol. 2019;68:505–508. doi: 10.1111/lam.13148. [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW, Tabor H, Xie QW. Spermidine synthase of Escherichia coli: localization of the speE gene. Proc Natl Acad Sci U S A. 1986;83:6040–6044. doi: 10.1073/pnas.83.16.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui Y, Saroj SD, Sakamoto A, Yoshida T, Higashi K, Kurihara S, Suzuki H, Toida T, Kashiwagi K, Igarashi K. Properties of putrescine uptake by PotFGHI and PuuP and their physiological significance in Escherichia coli. Amino Acids. 2014;46:661–670. doi: 10.1007/s00726-013-1517-x. [DOI] [PubMed] [Google Scholar]
- Thongbhubate K, Nakafuji Y, Matsuoka R, Kakegawa S, Suzuki H. Effect of spermidine on biofilm formation in Escherichia coli K-12. J Bacteriol. 2021;203:e00652–e720. doi: 10.1128/JB.00652-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gabain A, Bujard H. Interaction of E. coli RNA polymerase with promotors of coliphage T5. Mol Gen Genet. 1977;157:301–311. doi: 10.1007/BF00268667. [DOI] [PubMed] [Google Scholar]
- von Gabain A, Bujard H. Interaction of Escherichia coli RNA polymerase with promoters of several coliphage and plasmid DNAs. Proc Natl Acad Sci U S A. 1979;76:189–193. doi: 10.1073/pnas.76.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendisch VF, Mindt M, Pérez-García F. Biotechnological production of mono-and diamines using bacteria: recent progress, applications, and perspectives. Appl Microbiol Biotechnol. 2018;102:3583–3594. doi: 10.1007/s00253-018-8890-z. [DOI] [PubMed] [Google Scholar]
- Xu D, Zhang L. Metabolic engineering of Escherichia coli for agmatine production. Eng Life Sci. 2019;19:13–20. doi: 10.1002/elsc.201800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhang L. Increasing agmatine production in Escherichia coli through metabolic engineering. J Agric Food Chem. 2019;67:7908–7915. doi: 10.1021/acs.jafc.9b03038. [DOI] [PubMed] [Google Scholar]
- Yamanobe T, Kurihara Y, Uehara H, Komoto T. Structure and characterization of nylon 46. J Mol Struct. 2007;829:80–87. doi: 10.1016/j.molstruc.2006.06.010. [DOI] [Google Scholar]
- Yoshida M, Kashiwagi K, Shigemasa A, Taniguchi S, Yamamoto K, Makinoshima H, Ishihama A, Igarashi K. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem. 2004;279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers used for strain construction. Table S2. Primers used for plasmid construction. Table S3. Primers used for Realtime PCR. Fig. S1. Comparison of extracellular putrescine concentration per OD600 among strains of the Keio collection. M9 glucose medium (60 mL in 100-mL Erlenmeyer flask with stirrer bar) was inoculated with the pre-culture at the initial OD600 of 0.06. The flasks were set in the 7-L rectangular anaerobic jar with 2 AnaeroPack-Anaero sachets (Mitsubishi Gas Chemical; Tokyo, Japan) to form an anaerobic environment and the jar was set on the stirrer in the microbiological incubator. The culture medium was stirred at 150 rpm and kept at 37°C. After 10 h of incubation, the OD600 of the culture was measured and 0.4 mL of the culture was centrifuged. Thirty μL of 100% (w/v) TCA was mixed with 0.3 mL of the supernatant, filtrated through the membrane filter, and then subjected to HPLC analysis as described in the main text. Asterisks indicate the genes with significant differences in transcription levels. Fig. S2. Q100TAG mutation in the yifE gene promotes putrescine production. Extracellular concentration of putrescine of AI32 (pQE-80L/MG1655, open diamond) and AI33 (pQE-80L::yifEQ100TAG/MG1655, closed square) cultured at 37°C in minimal M9 supplemented with 0.2% glucose. 100 μg/mL of ampicillin was added to maintain the plasmids. When the OD600 reached 0.5, the 0.5 mM IPTG was added. Fig. S3. The presence of hdfR had a slight, if any, effect on the increase in extracellular putrescine concentration by YifEQ100TAG. (A) Cell growth and (B) extracellular concentration of putrescine of AI32 (pQE-80L/MG1655, open circle), AI33 (pQE-80L::yifEQ100TAG/MG1655, closed triangle), and KT218 (pQE-80L::yifEQ100TAG/ MG1655 but ΔhdfR::FRT-kanR-FRT, closed square) cultured at 37°C in LB supplemented with 100 μg/mL of ampicillin. When the OD600 reached 0.4, 0.02 mM IPTG was added.
Data Availability Statement
Some data are not shown in the text and supplement. However, they are available from the authors upon reasonable request. The strains and plasmids used in this work can be shared by exchanging MTA.