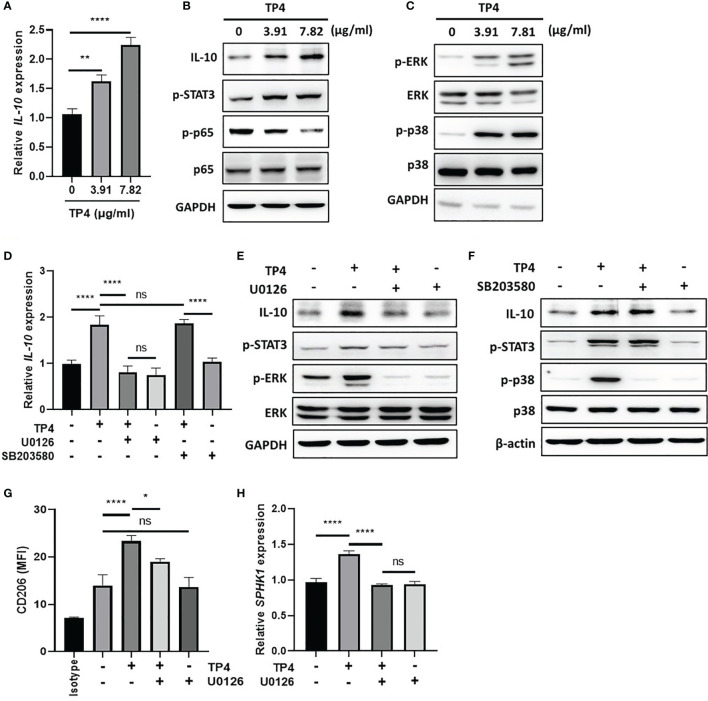
Figure 6.
TP4 induces IL-10 expression via MAPK/ERK pathway in THP-1/GV cells and promotes M2c phenotypes. (A) IL-10 expression was measured after 4 h TP4 (3.91 or 7.82 μg/ml) treatment of THP-1/GV cells by qRT-PCR. Data were represented the normalized target gene amount relative to the group of 0 μg/ml. (B) IL-10, and phosphorylated STAT3 (p-STAT3) and NFκB p65 subunit (p-p65) after 4 h TP4 (3.91 or 7.82 μg/ml) treatment of THP-1/GV cells were detected by immunoblotting. GAPDH was used as an internal control to show equal protein loading. (C) Detection of total and phosphorylated ERK (ERK/p-ERK) and p38 (p38/p-p38) after 4 h TP4 (3.91 or 7.82 μg/ml) treatment of THP-1/GV cells by immunoblotting. GAPDH was used as an internal control to show equal protein loading. (D) THP-1/GV cells were pre-treated with ERK inhibitor (U0126; 10 μM) or p38 inhibitor (SB203580; 10 μM) for 2 h in the serum-free medium. After incubation, TP4 (7.82 μg/ml) was treated for 4 h. IL-10 expression was measured by qRT-PCR. Data were represented the normalized target gene amount relative to the untreated control group. (E) Detection of IL-10, p-STAT3, p-ERK, and ERK after U0126 (10 μM) and TP4 (7.82 μg/ml) treatments in THP-1/GV cells by immunoblotting. GAPDH was used as an internal control to show equal protein loading. (F) Detection of IL-10, p-STAT3, p-p38 and p38 after SB203580 (10 μM) and TP4 (7.82 μg/ml) treatments in THP-1/GV cells by immunoblotting. GAPDH was used as the internal control to show equal protein loading. (G) Macrophage surface marker (CD206) was detected by flow cytometry; the mean fluorescence intensity (MFI) is shown. The black bar graph indicates isotype controls. (H) SPHK-1 expression after U0126 (10 μM) and TP4 (7.82 μg/ml) treatments in THP-1/GV cells were measured by qRT-PCR. Data were represented the normalized target gene amount relative to the untreated control group. Data are presented as mean ± SD of three independent experiments (*P < 0.05; **P < 0.01; ****P < 0.0001; ns, not statistically significant).