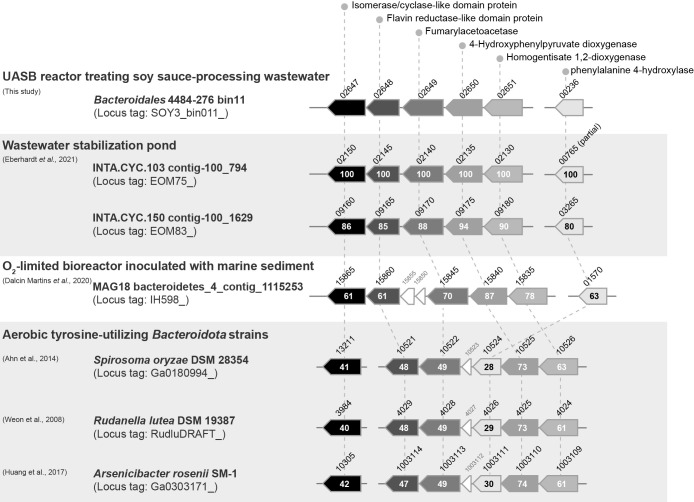
Fig. 3.
Comparison of gene cassettes associated with the bioconversion of 4-hydroxyphenylpyruvate to acetoacetate/fumarate found in uncultured Bacteroidales 4484-276 bin11, previously reported metagenomic contigs derived from probable microaerobic ecosystems (Eberhardt et al., 2020; Dalcin Martins et al., 2021), and aerobic Bacteroidota isolates (Weon et al., 2008; Ahn et al., 2014; Huang et al., 2017). An uncultured Bacteroidales 4484-276 bin11-associated cassette encodes homogentisate 1,2-dioxygenase, 4-hydroxyphenylpyruvate dioxygenase, fumarylacetoacetase, flavin reductase-like domain protein, and putative isomerase/cyclase-like domain protein. Phenylalanine 4-hydroxylase is also shown. Abbreviated locus tags are shown (e.g., ‘SOY3_bin011_02651’ as ‘02651’ in the row of Bacteroidales 4484-276 bin11). Numbers in gene boxes indicate amino acid sequence identity (%) to the corresponding gene of uncultured Bacteroidales 4484-276 bin11. The contigs INTA.CYC.103 contig-100_794 (37,009 bp), INTA.CYC.150 contig-100_1629 (24,086 bp), and MAG18 bacteroidetes_4_contig_1115253 (29,273 bp) were originally named as Candidatus Falkowbacteria bacterium (accession no. SABQ01000012), Clostridia bacterium (SABY01000055), and Bacteroidales bacterium (JACZJL010000162), respectively, in the NCBI database. We checked the phylogenetic assignment of these contigs by a blastp search (Altschul et al., 1990) for some functionally important enzymes; i.e., RNA polymerase (locus tag: EOM75_02175), DNA repair protein (EOM75_02190), and chromosomal replication initiator protein (EOM75_02085) for INTA.CYC.103 contig-100_794; RNA polymerase (EOM83_09135), DNA repair protein (EOM83_09120), and aminotransferase (EOM83_09145) for INTA.CYC.150 contig-100_1629; 5′/3′-nucleotidase (IH598_15930), transketolase (IH598_15890), and catalase (IH598_15870) for MAG18 bacteroidetes_4_contig_1115253. Since all enzymes are closely related to Bacteroidota organisms, we temporarily concluded that these contigs may be derived from Bacteroidota-related microorganisms.