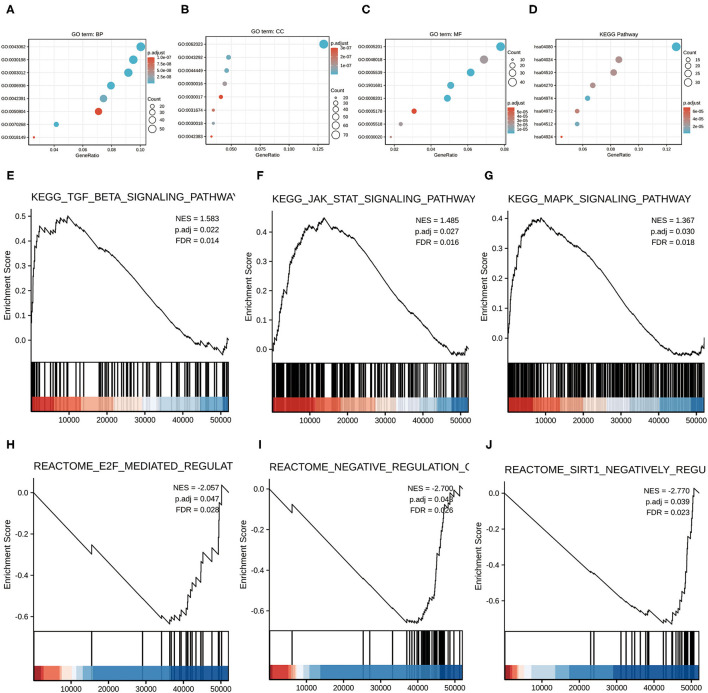
Figure 2.
Functional enrichment analysis of 256 differentially expressed genes (DEGs) between high and low expression of lncRNA MSC-AS1 in patients with gastric cancer in TCGA. (A) Enriched Gene Ontology (GO) terms in the biological process category. (B) Enriched GO terms in the cellular component category. (C) Enriched GO terms in the molecular function category. (D) Enriched GO terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) category. The X-axis represents the proportion of DEGs, and the Y-axis represents different categories. The different colors indicate different properties, and the different sizes represent the numbers of DEGs. (E–J) Enrichment plots from Gene Set Enrichment Analysis (GSEA). The TGF-β signaling pathway, the JAK-STAT signaling pathway, and the MAPK signaling pathway were differentially enriched in the long non-coding RNA (lncRNA) MSC-AS1 high-expression phenotype. In the lncRNA MSC-AS1 low-expression phenotype, enriched pathways included E2F mediated regulation of DNA replication, negative regulation of NOTCH4 signaling, and SIRT1 negatively regulates RNA expression.