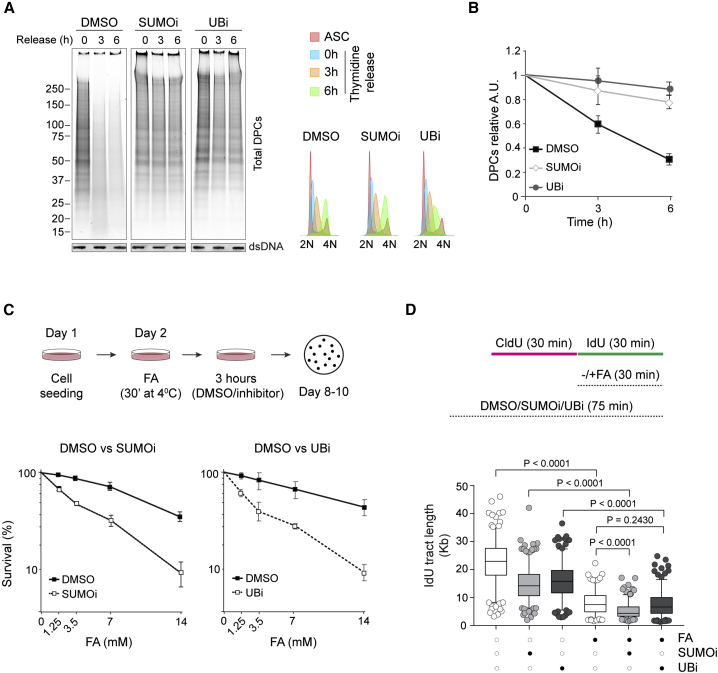
Figure 2.
SUMO and ubiquitin are required for replication-coupled DPC repair
(A) SUMOylation and ubiquitylation inhibition block DPC removal during S phase progression. HeLa cells were synchronized in G1/S with double thymidine block and released in the presence of DMSO, 25 μM 2-D08 (SUMOi), or 5 μM MLN7243 (UBi). Total DPCs were isolated by RADAR and detected by Flamingo protein gel staining. Slot blot with anti-dsDNA was used as a loading control. Right panel shows cell cycle distribution by fluorescence-activated cell sorting (FACS) analysis of the DNA content (propidium iodide).
(B) Quantification of DPC removal for the experiment in (A).
(C) SUMOylation and ubiquitylation inhibition sensitize cells to FA. Schematic of the survival assay protocol (upper panel). HeLa cells were exposed to the indicated concentrations of FA for 30 min at 4°C and let recover for 3 h in the presence of DMSO, 25 μM 2-D08 (SUMOi), or 5 μM MLN7243 (UBi). Colonies were allowed to grow for 8–10 days before fixation and counting (n = 2, mean ± SD).
(D) SUMOylation and ubiquitylation inhibition reduce DNA replication speed. Box and whiskers plot for DNA combing analysis. HEK293 cells were allowed to incorporate chloro-deoxyuridine (CldU) for 30 min and iodo-deoxyuridine (IdU) for an additional 30 min in the presence of DMSO, 50 μM 2-D08 (SUMOi), or 5 μM MLN7243 (UBi). Where indicated, 450 μM FA was added for the duration of IdU incubation. The length of IdU tracts was measured with FiberVision software, and statistical significance was calculated using an unpaired t test (Mann-Whitney) (170–260 events). The graph is representative of two independent experiments.