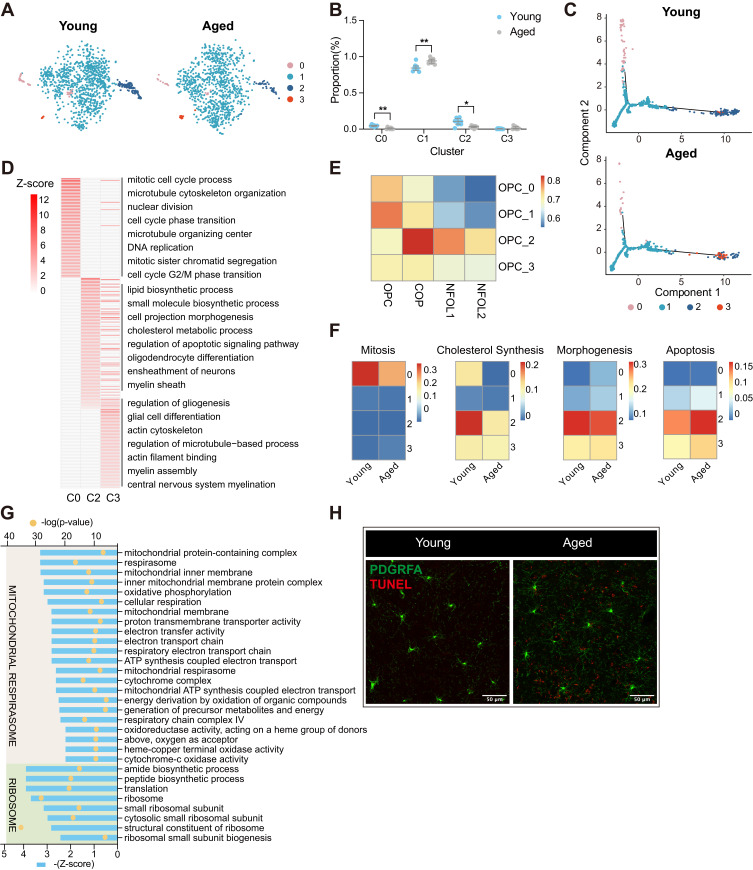
Figure 4.
Subgroup division and age-related transcriptomic alteration of OPCs (A) T-distributed stochastic neighbor embedding (t-SNE) projection of four OPC subclusters from young and aged mice. (B) Dot plot showing the proportion of each subcluster in young and aged OPCs (**P-value < 0.01, *P-value < 0.05 by Wilcoxon Rank Sum test, n = 8 samples per condition). (C) Cellular trajectory of all OPC subclusters, generated by Monocle DDRTree dimensionality reduction algorithm. (D) Heatmap showing part of significant (P-value < 0.05) GO enrichment terms using DEGs between the C0, C2, C3 and C1 subset of OPCs. Numbers in legend represented Z-score, positive values indicated upregulation. (E) Heatmap of Pearson correlation between OPCs subclusters with the related populations mentioned by Marques et al (F) Heatmaps depicting the average gene expression on mitosis, cholesterol synthesis, morphogenesis and apoptosis regulation module in four subclusters from young and aged OPCs, respectively. (G) Plot showing part of GO terms predicted to be inhibited significantly (Z-score ≤ −2, P-value < 0.05) along with aging in OPC C1. These terms were divided into two functional clusters (ribosome and mitochondrial respirasome) according to the biologic functions they were involved in. (H) Representative confocal microscopic images in young and old cortex staining for PDGFRA and TUNEL. Scale bar: 50μm.
Abbreviations: OPCs, oligodendrocyte precursor cells; COP, committed OPC; NFOL, newly formed oligodendrocyte; DEGs, differentially expressed genes; GO, gene oncology.