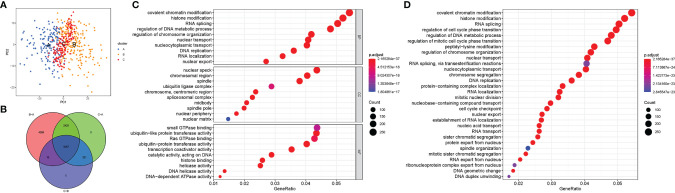
Figure 3.
The differential expression of genes in each cluster and the function analysis of co-expressed genes among three clusters. (A) Principal component analysis for the transcriptome profiles of cluster A, B, and C; (B) Venn diagram showing the co-expressed genes in the three clusters; (C) GO analysis of the co-expressed genes in three clusters; (D) KEGG analysis of the co-expressed genes in three clusters and the result showed that the co-expressed genes were mainly enriched in pathways related to histone, cell cycle, DNA, and RNA functions.