Supplemental Digital Content is Available in the Text.
Mendelian randomization study supports that depression is a causal risk factor for headache and pain at neck/shoulder, back, and abdominal/stomach.
Keywords: Depression, Pain, Mendelian randomization study, Functional annotation analysis, Causal inference, Genetics
Abstract
To understand a putative causal link for depression and pain, we retrieved summary statistics from genome-wide association studies conducted for pain at 7 different body sites (N = 151,922-226,683) and major depression disorder (MDD, Ncase/control = 246,363/561,190). We conducted a bidirectional Mendelian randomization analysis using distinct genome-wide association studies-identified single nucleotide polymorphisms for each trait as instrumental variables and performed several sensitivity analyses to verify Mendelian randomization assumptions. We also conducted functional annotation analysis using 396 tissue-specific annotations from the roadmap project. Across 7 different body sites, genetic predisposition to depression was associated with pain at the neck/shoulder (odds ratio [OR] = 1.08 per one log-unit increase in depression risk, 95% confidence interval [CI]: 1.06-1.10), back (OR = 1.05, 95% CI: 1.04-1.07), abdominal/stomach (OR = 1.03, 95% CI: 1.02-1.04), as well as headache (OR = 1.10, 95% CI: 1.07-1.12), but not with pain on the face, hip, and knee. In the reverse direction, genetically instrumented multisite chronic pain (OR = 1.78 per one increment in the number of pain site, 95% CI: 1.51-2.11) and headache (OR = 1.55 per one log-unit increase in headache risk, 95% CI = 1.13-2.10) were associated with MDD. Functional annotation analysis showed differential clustering patterns where depression clustered closely with headache and neck/shoulder pain, exhibiting substantial brain tissue enrichment. Our study indicates that depression is a causal risk factor for headache and pain localized at neck/shoulder, back, and abdominal/stomach, rather than pain at face, hip, and knee, and suggests common neurological pathologies underlying the development of depression, headache, and neck/shoulder pain.
1. Introduction
Depression and pain are 2 common and deleterious disorders that cause substantial economic and societal burden. Clinical observations have long recognized the comorbidity and interaction between depression and pain, where both conditions often coexist, respond to similar treatments, aggravate each other, and share common biological mechanisms.20
Despite a highly heterogeneous and chronic nature of depression and pain, both traits exhibit a significant genetic component in its development. For example, a recent study has meta-analyzed 807,553 individuals (246,363 cases and 561,190 controls) from the 3 largest genome-wide association studies (GWAS) of depression and identified 102 independent variants, 269 genes, and 15 gene-sets associated with major depression disorder (MDD), including genes and pathways associated with synaptic structure and neurotransmission.14 Similarly, the polygenic architecture of pain has been elucidated by a GWAS of multisite chronic pain (MCP) conducted in ∼380,000 UK Biobank participants, which identified independent lead single-nucleotide polymorphisms (SNPs) at 39 risk loci.16 Furthermore, site-specific GWAS(s) have been performed for pain at different body parts including neck/shoulder,25 back,30 head,22 and knee23 and identified GWAS-significant genetic variants ranging from 3 loci (associated with back, knee, or neck/shoulder pain) to 28 loci (associated with headache).
The relationship between depression and pain has been studied to some extent, yet the results remain uncertain. A recent genetic correlation analysis has identified positive and significant shared genetic basis of MDD with headache ( = 0.39), neck/shoulder pain ( = 0.40), stomach/abdominal pain ( = 0.53), and back pain ( = 0.36), but not with facial, hip, or knee pain (all close to 0).24 These estimates can reflect pleiotropy, where specific genetic alleles increase risk to both phenotypes, but it can also reflect a directional and/or causal association. The latter can be examined by Mendelian randomization (MR) design, which uses genetic variants as instrumental variables (IVs) and makes causal inference. So far, only 2 MR studies have been performed to explore a putative causal relationship regarding depression and pain, both in a format of secondary or complementary analysis. One study did not find any causal relationship between MDD and headache,24 whereas the other identified a causal effect between MDD and MCP in both directions.16
As additional MDD-associated loci have been discovered and novel statistical approaches have been developed, we aim to update and extend previous findings by conducting a bidirectional MR, leveraging summary statistics of the largest GWAS conducted in MDD and localized pain at 7 different body sites. We complement our bidirectional MR with a cell-type specific functional annotation analysis to partition the heritability and to understand the shared genetic origin across traits.
2. Materials and methods
We conduct the current MR study applying a standard two-sample framework where the IV-exposure and IV-outcome associations are from 2 GWAS (Supplementary Fig. 1, available at http://links.lww.com/PAIN/B367).
2.1. Major depression disorder genome-wide association studies
The largest GWAS of MDD was conducted meta-analyzing data on 246,363 cases of self-reported clinical diagnosis of depression or self-reported broad depression and 561,190 controls, all of European ancestry.14 A total of 102 independent variants (biallelic common SNPs, P<5 × 10−8) were identified. We included these 102 SNPs as our IVs(MDD) and extracted IV(MDD)-MDD associations (beta-coefficients, standard errors) from the MDD GWAS (Supplementary Table 1, available at http://links.lww.com/PAIN/B367).
Noteworthily, this MDD GWAS contained UK Biobank participants, which overlap to some extent (28%) with the localized pain GWAS. We therefore additionally extracted 44 MDD-associated SNPs identified by an earlier GWAS comprising 135,458 cases and 344,901 controls, all of European ancestry, to conduct a sensitivity analysis.32 There the GWAS contains a small proportion of participants from UK Biobank (N = 29,740, 6%), resulting in a minimal overlap with the pain GWAS.
2.2. Pain genome-wide association studies
Large-scale GWAS(s) on pain by different body sites were conducted analyzing the UK Biobank participants (N = 151,922-226,683). Information was collected through a specific pain-related questionnaire, which included a question “In the last month have you experienced any of the following that interfered with your usual activities?”. The options were: (1) headache; (2) facial pain; (3) neck or shoulder pain; (4) back pain; (5) stomach or abdominal pain; (6) hip pain; (7) knee pain; (8) pain all over the body; (9) none of the above; (10) prefer not to say.24 Here, we included pain at 7 specific sites, namely headache, neck/shoulder, back, abdominal/stomach, facial, knee, and hip. For each pain phenotype, cases were defined as those who selected a specific pain site option, regardless of whether they had selected other options. Controls were those who selected the “none of the above” option. We did not include “pain all over the body” because this measure, taken as a proxy for chronic widespread pain, represents a different clinical syndrome from localized chronic pain and does not necessarily directly reflect chronic pain at 7 body sites. Alternatively, we obtained data from a large GWAS on MCP with 387,649 UK Biobank participants, all of European ancestry.16 Multisite chronic pain was measured as the number of body sites (from 0 to 7 sites) at which pain had lasted for at least 3 months.
We extracted the IV(MDD)-pain associations (beta-coefficients, standard errors) from each of the 7 pain GWAS(s) (Supplementary Table 1, available at http://links.lww.com/PAIN/B367). We also retrieved the full-set GWAS summary data for functional annotation analysis.
Because depression and pain often co-occur, we next tried to explore whether pain affects depression onset (opposite to the aforementioned depression to pain relationship). For most site-specific pain, for example, knee, back, shoulder/neck, less than 3 GWAS-significant SNPs were discovered, making the analysis underpowered. We therefore included only 2 pain phenotypes with better power: headache defined as self-reported broad-sense headache with 28 independently associated SNPs (Supplementary Table 2, available at http://links.lww.com/PAIN/B367) and MCP with 39 independently associated SNPs (Supplementary Table 3, http://links.lww.com/PAIN/B367).
2.3. Lifestyle factors genome-wide association studies
Pain, as an objective assessment, is highly heterogeneous and influenced by other factors. We therefore incorporated summary statistics from the GWAS of several heritable lifestyle exposures, including smoking,21 alcohol consumption,21 physical activity,8 educational attainment,19 and body mass index (BMI).33 We first tested the association between each localized pain and the 5 lifestyle factors, and then included IVs associated with these lifestyle factors in a multivariable MR (MVMR) analysis to adjust for their effects.
Characteristics of all GWAS data are summarized in Supplementary Table 4 (available at http://links.lww.com/PAIN/B367). The GWAS summary data do not contain any personal information, and the original GWAS have obtained ethical approval from relevant ethics review committees.
2.4. Statistical analysis
2.4.1. Mendelian randomization analysis
We first evaluated a bidirectional causal relationship between depression and pain, applying several MR approaches including a random-effect inverse variance-weighted method (IVW),6 a weighted median approach,4 and an MR-Egger regression.3
Briefly, the random-effect IVW pools estimate from each IV and provides causal estimation assuming all IVs are valid or are invalid in a way that overall pleiotropy is balanced to be zero. Complementary to IVW, we also used a weighted median approach, which provides consistent estimates even when up to 50% of the analyzed genetic variants are invalid IVs. Finally, we performed MR-Egger regression to test for bias due to directional pleiotropy, where the average of direct effects of the tested genetic variants on outcome is nonzero. The heterogeneity between IVs was tested using Cochran Q test.
Three important model assumptions need to be satisfied for MR to yield unbiased causal estimates. Namely, IVs should be robustly associated with the exposure (relevance), affect outcome only through the exposure (exclusion restriction), and should not be associated with confounders in the exposure–outcome relationship (exchangeability). We therefore performed important sensitivity analyses to validify model assumptions. For example, we excluded palindromic IVs or IVs that were associated with potential confounding traits according to GWAS catalog (https://www.ebi.ac.uk/gwas/). We also used an MVMR approach to adjust for potential horizontal pleiotropy acting through confounders such as smoking, alcohol consumption, BMI, and levels of education.7 All MR analyses were conducted using the “TwoSampleMR” package in R software version 3.6.0.11 The causal estimates with binary exposures, including MDD and localized pain, represent the change in the outcome per unit change in the exposure on the log odds scale, whereas the causal estimates for multisite chronic pain, a numeric exposure ranging from 0 to 7 pain sites, represent the change in the outcome per one increment in the number of pain sites.
2.4.2. Functional annotation analysis
To understand the (dis)similarity across traits, we further partitioned heritability using stratified-LD score regression leveraging genome-wide genetic variants of 7 localized pain and MDD.9 This method partitions SNPs into functional categories and calculates category-specific enrichment based on the assumption that a category of SNPs is enriched for heritability if SNPs with high linkage disequilibrium to that category have higher χ2 statistics than SNPs with low linkage disequilibrium to that category. Details of our functional annotation analysis are presented in Supplementary Note 1 (available at http://links.lww.com/PAIN/B367).
We further divided those 396 cell-type–specific annotations into 9 broad groups (adipose, central nervous system (CNS), digestive system, cardiovascular, musculoskeletal and connective tissue, immune and blood, liver, pancreas, and other) by taking a union of the cell-type–specific annotations within each group (eg, SNPs with any of the 6 histone modifications in any hematopoietic and immune cells were considered as one big category). All functional annotation analyses were conducted using the LD score regression software,5,9 and enrichment values were transformed into color scale and visualized by hierarchical clustering.
To account for multiple comparisons, we considered a P-value smaller than 0.05 as suggestive significance; a Bonferroni-corrected P-value was applied based on the specific numbers of comparisons made in each analysis.
3. Results
As shown in Table 1, across 7 different body sites, genetic predisposition to depression was associated with pain on the neck/shoulder (odds ratio [OR] = 1.08, 95% confidence interval [CI]: 1.06-1.10, P = 6.5 × 10−17), back (OR = 1.05, 95% CI: 1.04-1.07, P = 8.5 × 10−11), abdominal/stomach (OR = 1.03, 95% CI: 1.02-1.04, P = 2.1 × 10−7), as well as headache (OR = 1.10, 95% CI: 1.07-1.12, P = 7.3 × 10−15). All P-values passed Bonferroni-corrected threshold using the IVW method (P < 0.05/7). The results remained consistent in both magnitude and direction using the weighted median approach. We did not observe any apparent horizontal pleiotropy as indicated by MR-Egger intercepts where all P-values for intercepts were greater than 0.05. Because MR-Egger regression produces twice as large standard errors as that of IVW, as expected, confidence intervals from MR-Egger were slightly inflated. Noteworthily, the effect of depression on back pain attenuated to null in the MR-Egger regression (OR = 0.99, P = 0.78). On the contrary, we did not observe any significant effect of depression with pain on face (OR = 1.00, P = 0.07), hip (OR = 1.01, P = 0.06), or knee (OR = 0.99, P = 0.10).
Table 1.
Genetic predisposition to major depression disorder and risk of pain: the results from Mendelian randomization analysis.
| Methods | # SNP | OR (95% CI) | P | P for intercept or heterogeneity | # SNP | OR (95% CI) | P | P for intercept or heterogeneity | # SNP | OR (95% CI) | P | P for intercept or heterogeneity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full set | Remove SNPs associated with confounding traits | Remove palindromic SNPs | ||||||||||
| Multisite chronic pain | ||||||||||||
| IVW | 102 | 1.28 (1.23-1.34) | 2.9 × 10 −27 | < 0.001 | 76 | 1.26 (1.20-1.33) | 1.1 × 10 −18 | < 0.001 | 89 | 1.27 (1.21-1.33) | 5.6 × 10 −22 | < 0.001 |
| MR-Egger | 102 | 1.14 (0.95-1.37) | 0.16 | 0.21 | 76 | 1.07 (0.87-1.32) | 0.53 | 0.11 | 89 | 1.13 (0.93-1.38) | 0.21 | 0.24 |
| Weighted median | 102 | 1.19 (1.14-1.24) | 2.5 × 10 −14 | 76 | 1.18 (1.12-1.25) | 1.3 × 10 −9 | 89 | 1.18 (1.12-1.24) | 1.0 × 10 −10 | |||
| Headache | ||||||||||||
| IVW | 95 | 1.10 (1.07-1.12) | 7.3 × 10 −15 | < 0.001 | 69 | 1.07 (1.05-1.10) | 1.1 × 10 −7 | < 0.001 | 83 | 1.09 (1.06-1.12) | 2.3 × 10 −11 | < 0.001 |
| MR-Egger | 95 | 1.06 (0.96-1.16) | 0.26 | 0.44 | 69 | 1.00 (0.90-1.11) | 0.95 | 0.16 | 83 | 1.07 (0.96-1.18) | 0.21 | 0.68 |
| Weighted median | 95 | 1.11 (1.08-1.13) | 5.9 × 10 −15 | 69 | 1.06 (1.03-1.10) | 5.7 × 10 −5 | 83 | 1.09 (1.06-1.12) | 2.3 × 10 −9 | |||
| Neck/shoulder pain | ||||||||||||
| IVW | 102 | 1.08 (1.06-1.10) | 6.5 × 10 −17 | < 0.001 | 76 | 1.07 (1.05-1.09) | 3.6 × 10−10 | 0.002 | 89 | 1.07 (1.05-1.10) | 1.6 × 10 −13 | < 0.001 |
| MR-Egger | 102 | 1.04 (0.96-1.12) | 0.35 | 0.27 | 76 | 1.03 (0.95-1.12) | 0.48 | 0.40 | 89 | 1.04 (0.97-1.13) | 0.28 | 0.44 |
| Weighted median | 102 | 1.06 (1.04-1.09) | 4.6 × 10 −9 | 76 | 1.05 (1.03-1.08) | 9.0 × 10 −5 | 89 | 1.06 (1.03-1.08) | 2.8 × 10 −6 | |||
| Back pain | ||||||||||||
| IVW | 95 | 1.05 (1.04-1.07) | 8.5 × 10 −11 | 0.03 | 69 | 1.06 (1.03-1.08) | 2.1 × 10 −7 | 0.01 | 83 | 1.05 (1.03-1.07) | 3.5 × 10 −8 | 0.02 |
| MR-Egger | 95 | 0.99 (0.93-1.06) | 0.78 | 0.06 | 69 | 0.97 (0.90-1.06) | 0.54 | 0.05 | 83 | 0.99 (0.93-1.06) | 0.77 | 0.09 |
| Weighted median | 95 | 1.05 (1.03-1.07) | 2.3 × 10 −5 | 69 | 1.05 (1.02-1.08) | 4.5 × 10 −4 | 83 | 1.04 (1.02-1.07) | 3.3 × 10 −4 | |||
| Abdominal/stomach pain | ||||||||||||
| IVW | 102 | 1.03 (1.02-1.04) | 2.1 × 10 −7 | 0.005 | 76 | 1.02 (1.01-1.03) | 7.5 × 10 −5 | 0.05 | 89 | 1.02 (1.01-1.03) | 4.5 × 10 −6 | 0.01 |
| MR-Egger | 102 | 1.02 (0.98-1.06) | 0.38 | 0.71 | 76 | 1.00 (0.96-1.05) | 0.84 | 0.43 | 89 | 1.02 (0.98-1.06) | 0.29 | 0.95 |
| Weighted median | 102 | 1.03 (1.01-1.04) | 9.4 × 10 −5 | 76 | 1.02 (1.01-1.03) | 2.2 × 10 −3 | 89 | 1.02 (1.01-1.04) | 1.1 × 10 −3 | |||
| Facial pain | ||||||||||||
| IVW | 102 | 1.00 (0.99-1.01) | 0.07 | 0.64 | 76 | 1.00 (0.99-1.01) | 0.15 | 0.43 | 89 | 1.00 (0.99-1.01) | 0.31 | 0.66 |
| MR-Egger | 102 | 1.00 (0.98-1.02) | 0.73 | 0.43 | 76 | 0.99 (0.97-1.02) | 0.51 | 0.30 | 89 | 1.00 (0.98-1.02) | 0.79 | 0.98 |
| Weighted median | 102 | 1.00 (0.99-1.01) | 0.27 | 76 | 1.00 (0.99-1.01) | 0.40 | 89 | 1.00 (0.99-1.01) | 0.31 | |||
| Hip pain | ||||||||||||
| IVW | 102 | 1.01 (0.99-1.02) | 0.06 | 0.77 | 76 | 1.01 (1.00-1.02) | 0.02 | 0.72 | 89 | 1.01 (1.00-1.02) | 0.03 | 0.96 |
| MR-Egger | 102 | 0.99 (0.96-1.03) | 0.69 | 0.39 | 76 | 1.02 (0.97-1.06) | 0.48 | 0.86 | 89 | 1.00 (0.96-1.04) | 0.91 | 0.54 |
| Weighted median | 102 | 1.00 (0.99-1.02) | 0.67 | 76 | 1.01 (0.99-1.02) | 0.48 | 89 | 1.01 (0.99-1.02) | 0.45 | |||
| Knee pain | ||||||||||||
| IVW | 102 | 0.99 (0.98-1.01) | 0.10 | 0.01 | 76 | 0.99 (0.98-1.01) | 0.23 | 0.07 | 89 | 0.99 (0.97-1.00) | 0.04 | 0.02 |
| MR-Egger | 102 | 0.98 (0.93-1.03) | 0.46 | 0.71 | 76 | 0.98 (0.92-1.04) | 0.50 | 0.69 | 89 | 0.98 (0.93-1.04) | 0.55 | 0.94 |
| Weighted median | 102 | 0.98 (0.96-1.00) | 0.02 | 76 | 0.98 (0.96-1.00) | 0.04 | 89 | 0.98 (0.96-0.99) | 0.01 | |||
CI, confidence interval; IVW, inverse variance-weighted; MR, Mendelian randomization; OR, odds ratio; SNP, single-nucleotide polymorphisms.
Sensitivity analysis removing palindromic IVs (Table 1) revealed similar findings on that genetically predicted depression increased sensitivity of pain on head, neck/shoulder, back, as well as abdominal/stomach, but not pain on face, hip, or knee. Consistent findings were observed excluding IVs associated with important potential confounders in genome-wide significance as revealed by the GWAS catalog (Table 1) as well as using 44 MDD-associated IVs identified in an earlier GWAS possessing negligible sample overlap with pain GWAS (Supplementary Table 5, available at http://links.lww.com/PAIN/B367). These results corroborated each other and support the robustness of our primary findings.
Among 5 lifestyle exposures, genetically predicted BMI, smoking, education, and alcohol consumption were significantly associated with at least one localized pain after Bonferroni-correction (P < 0.05/35 = 0.0014, Fig. 1 and Supplementary Table 6, available at http://links.lww.com/PAIN/B367). Adjusting for all confounders simultaneously in one multivariable MR model did not alter our results (headache: OR = 1.08, P = 3.2 × 10−40; neck/shoulder pain: OR = 1.05, P = 6.8 × 10−17; back pain: OR = 1.03, P = 2.2 × 10−9; abdominal/stomach pain: OR = 1.02, P = 7.4 × 10−18). No significant association was observed for facial, hip, or knee pain. Adjusting for each confounder sequentially revealed similar findings (Table 2).
Figure 1.
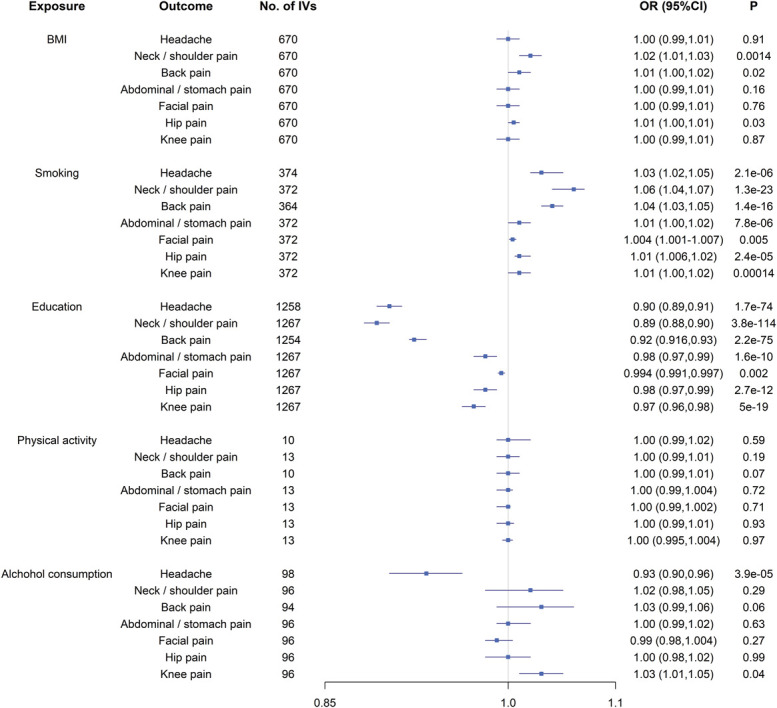
The putative causal effect of known confounding factors on pain: the results from Mendelian randomization analysis using the IVW approach. Squares and horizontal bars represent the odds ratios and confidence intervals of factor with the risk of pain, respectively. BMI, body mass index; CI, confidence interval; IVW, inverse-variance weighted approach; OR, odds ratio.
Table 2.
Theresults from multivariable Mendelian randomization analysis adjusting for confounders.
| Methods | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | xOR (95% CI) | P | OR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adjust for all | Adjust for BMI | Adjust for smoking | Adjust for education | Adjust for alcohol | Adjust for exercise | |||||||
| MDD on pain | ||||||||||||
| Multisite chronic pain | 1.16 (1.14-1.19) | 3.1 × 10 −39 | 1.20 (1.17-1.24) | 8.0 × 10 −31 | 1.19 (1.15-1.22) | 4.9 × 10 −29 | 1.20 (1.17-1.22) | 1.2 × 10 −65 | 1.22 (1.17-1.26) | 1.4 × 10 −28 | 1.22 (1.18-1.27) | 4.4 × 10 −25 |
| Headache | 1.08 (1.07-1.10) | 3.2 × 10 −40 | 1.09 (1.07-1.11) | 5.4 × 10 −25 | 1.08 (1.06-1.10) | 1.6 × 10 −21 | 1.08 (1.07-1.09) | 1.4 × 10 −41 | 1.08 (1.06-1.10) | 6.9 × 10 −20 | 1.08 (1.06-1.10) | 8.3 × 10 −15 |
| Neck/shoulder pain | 1.05 (1.04-1.06) | 6.8 × 10 −17 | 1.07 (1.05-1.08) | 4.4 × 10 −18 | 1.05 (1.04-1.07) | 4.8 × 10 −15 | 1.06 (1.05-1.07) | 2.6 × 10 −32 | 1.06 (1.05-1.08) | 9.5 × 10 −17 | 1.07 (1.05-1.08) | 2.3 × 10 −17 |
| Back pain | 1.03 (1.02-1.04) | 2.2 × 10 −9 | 1.04 (1.02-1.05) | 7.5 × 10 −8 | 1.04 (1.02-1.05) | 1.8 × 10 −9 | 1.04 (1.03-1.05) | 2.0 × 10 −18 | 1.04 (1.03-1.06) | 9.4 × 10 −11 | 1.05 (1.03-1.06) | 2.5 × 10 −12 |
| Abdominal/stomach pain | 1.02 (1.01-1.03) | 7.4 × 10 −18 | 1.02 (1.01-1.03) | 1.7 × 10 −10 | 1.02 (1.01-1.03) | 8.1 × 10 −11 | 1.02 (1.01-1.03) | 2.0 × 10 −18 | 1.02 (1.01-1.03) | 1.1 × 10 −10 | 1.02 (1.01-1.03) | 4.1 × 10 −8 |
| Facial pain | 1.004 (1.001-1.007) | 0.01 | 1.005 (1.001-1.009) | 0.01 | 1.004 (1.00-1.01) | 0.03 | 1.01 (1.00-1.02) | 3.3 × 10 −5 | 1.005 (1.001-1.009) | 0.01 | 1.004 (1.00-1.008) | 0.05 |
| Hip pain | 1.00 (0.99-1.01) | 0.45 | 1.00 (0.99-1.01) | 0.22 | 1.00 (0.99-1.01) | 0.37 | 1.006 (1.00-1.01) | 0.01 | 1.00 (0.99-1.01) | 0.11 | 1.00 (0.99-1.01) | 0.10 |
| Knee pain | 1.00 (0.99-1.01) | 0.87 | 1.01 (0.99-1.02) | 0.31 | 0.99 (0.98-1.00) | 0.13 | 1.00 (0.99-1.01) | 0.90 | 1.00 (0.98-1.004) | 0.27 | 0.99 (0.98-1.00) | 0.19 |
| Pain on MDD | ||||||||||||
| Multisite chronic pain | 1.83 (1.57-2.13) | 9.9 × 10 −15 | 1.82 (1.61-2.06) | 1.6 × 10 −21 | 1.73 (1.52-1.98) | 5.5 × 10 −16 | 1.78 (1.51-2.10) | 7.5 × 10 −12 | 1.87 (1.71-2.04) | 2.1 × 10 −43 | 1.42 (1.14-1.76) | 1.5 × 10 −3 |
| Headache | 2.62 (2.22-3.10) | 1.16 × 10 −29 | 3.04 (2.58-3.59) | 6.4 × 10 −40 | 2.61 (2.31-2.95) | 7.7 × 10 −54 | 1.17 (1.10-1.24) | 1.5 × 10 −7 | 3.32 (2.92-3.77) | 9.1 × 10 −76 | 1.23 (1.16-1.31) | 1.4 × 10 −11 |
Perhaps not surprisingly, our reverse directional MR identified a significant causal association of pain with depression (Table 3). We found that genetic predisposition to multisite chronic pain was associated with the risk of depression, and the results were consistent across different sets of IVs (all IVs: OR = 1.78, P = 1.6 × 10−11; removing palindromic IVs: OR = 1.78, P = 2.4 × 10−10; removing pleiotropic IVs: OR = 1.67, P = 8.9 × 10−9), statistical methods (weighted median: OR = 1.48, P = 8.3 × 10−7), or adjustment for known confounding lifestyle factors (Table 2, adjusted for all lifestyle factors: OR = 1.83, P = 9.9 × 10−15). Due to limited availability of headache-associated IVs, estimates of headache with depression displayed larger statistical uncertainty and directional inconsistency (Table 3, all IVs: IVW, OR = 1.55, 95% CI, 1.13-2.10, P = 0.005; MR-Egger, OR = 0.77, 96% CI, 0.30-1.98, P = 0.59; weighted median, OR = 1.42, 95% CI, 1.07-1.88, P = 0.01). No substantial pleiotropy was detected by MR-Egger intercept (P for intercept =0.14).
Table 3.
Genetic predisposition to pain and risk of major depression disorder: the results from Mendelian randomization analysis.
| Methods | # SNP | OR (95% CI) | P | P for intercept or heterogeneity | # SNP | OR (95% CI) | P | P for intercept or heterogeneity | # SNP | OR (95% CI) | P | P for intercept or heterogeneity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full set | Remove SNPs associated with confounding traits | Remove palindromic SNPs | ||||||||||
| Multisite chronic pain on MDD | ||||||||||||
| IVW | 37 | 1.78 (1.51-2.11) | 1.6 × 10 −11 | < 0.001 | 26 | 1.67 (1.40-1.98) | 8.9 × 10 −9 | < 0.001 | 34 | 1.78 (1.49-2.12) | 2.4 × 10 −10 | < 0.001 |
| MR-Egger | 37 | 1.60 (0.68-3.79) | 0.28 | 0.81 | 26 | 2.28 (0.88-5.94) | 0.19 | 0.52 | 34 | 1.57 (0.65-3.80) | 0.32 | 0.78 |
| Weighted median | 37 | 1.48 (1.27-1.73) | 8.3 × 10 −7 | 26 | 1.48 (1.22-1.79) | 5.4 × 10 −5 | 34 | 1.48 (1.26-1.74) | 1.9 × 10 −6 | |||
| Headache on MDD | ||||||||||||
| IVW | 26 | 1.55 (1.13-2.10) | 0.005 | < 0.001 | 20 | 1.77 (1.18-2.66) | 0.005 | < 0.001 | 23 | 1.35 (1.06-1.70) | 0.01 | 0.02 |
| MR-Egger | 26 | 0.77 (0.30-1.98) | 0.59 | 0.14 | 20 | 0.59 (0.16-2.16) | 0.44 | 0.10 | 23 | 0.77 (0.39-1.53) | 0.47 | 0.11 |
| Weighted median | 26 | 1.42 (1.07-1.88) | 0.01 | 20 | 1.77 (1.25-2.49) | 0.001 | 23 | 1.41 (1.07-1.85) | 0.01 | |||
CI, confidence interval; IVW, inverse variance-weighted; MDD, major depression disorder; MR, Mendelian randomization; OR, odds ratio.
Because depression and pain showed a strong genetic component (SNP-heritability), we further partitioned such heritability by cell-type–specific annotations. As presented in Supplementary Figure 2 and Supplementary Table 7 (available at http://links.lww.com/PAIN/B367), different clustering patterns were observed comparing cell-type–specific enrichment for depression with 7 localized pain. At 5 of the 6 chromatin marks, especially in 3 enhancer-related marks that are suggested to be more informative for tissue-specific disease enrichment (H3K27ac, H3K9ac, and H3K4me1), depression clustered closely with headache and neck/shoulder pain and exhibited substantial brain tissue components.
4. Discussion
To the best of our knowledge, this is the first comprehensive genetic analysis that systemically interrogates a causal relationship between depression and pain, 2 highly complicated and entangled disorders. We found evidence supporting for a connection between depression and pain at specific body sites such as head, neck/shoulder, back, and abdominal/stomach, but not at other sites including face, hip, or knee. We further discovered a brain tissue enrichment in depression, neck/shoulder pain, and headache, suggesting a neural mechanism underlying the causal link.
Our findings are in line with a previous report that has quantified the genetic correlation between depression and pain and found significantly shared genetic architecture of depression with headache ( = 0.39), neck/shoulder pain ( = 0.40), back pain ( = 0.36), and abdominal/stomach pain (=0.53), but not with facial, hip, or knee pain (all close to 0).24 Consistent with these results, we identified that genetic predisposition to depression targeted pain on certain body areas (head, neck/shoulder, back, and abdominal/stomach) rather than others (face, hip, and knee). The negative findings regarding facial, hip, or knee pain contradict observations from conventional epidemiological investigations. For example, a cohort study has followed 3006 patients and reported that depression increased the risk of temporomandibular disorders, a subgroup of facial pain problem (RR = 2.1, 95% CI = 1.5-3.0, P = 0.001).17 Another cohort study that has followed 3407 patients with osteoarthritis for 2 years found that depression was significantly associated with knee pain worsening.27 Moreover, a cross-sectional study of 2515 adult participants has suggested that elevated depression scores were significantly and independently associated with disabling chronic hip pain.29 Although genetic factors are likely to contribute to pain at different sites, our negative findings suggest that nongenetic triggers may be more relevant in the co-occurrence of facial, knee, or hip pain with depression. However, although our results suggest a strong evidence of null association of depression with facial, hip, and knee pain (ORs virtually equal to 1), we had limited power for these 3 pain traits due to small sample sizes. Therefore, our results need to be confirmed when larger GWAS data become available.
For those 4 body areas (head, neck/shoulder, back, and abdominal/stomach) where we have identified significant causal associations, the nature of such relationship remains uncertain with 2 potential explanations—a true causal relationship or confounding by pleiotropic factors. We attempted to reduce the likelihood of confounding through statistical approaches such as using curated IVs without pleiotropic effects, MR-Egger regression, and MVMR. The highly consistent results between MVMR adjusting for different covariates and our main analysis reflect the effect of depression on pain independent of potential confounding factors including smoking, alcohol intake, physical activity, educational attainment, and obesity, and careful analyses into additional confounders are warranted.
We further discovered that genetically predicted multisite chronic pain also increased the risk of depression. Such bidirectional relationship corroborates clinical observations as well as animal behavioral experiments on a reciprocal interaction between pain and depression, where depressive-like conditions exacerbate pain perception and the presence of chronic pain aggravates depressive-like behaviors.20 Indeed, prospective studies have suggested a reciprocal association between depression and headache where headache increased the likelihood of incident depression (RR = 1.44, 95% CI = 1.32-1.56)28 and depression was significantly associated with recurrent headache (OR = 1.6, 95% CI = 1.2-2.1, P = 0.001).2 Our result regarding genetic predisposition to headache and risk of depression needs to be validated by future studies due to its limited number of IVs and large statistical uncertainty.
Generally, the present MR study provides evidence supporting a putative causal relationship between depression and pain. The main etiological hypothesis is that these 2 disorders are linked by common underlying neurobiological mechanisms. At the tissue level, the functional brain reorganization in depression and pain has been investigated by a multitude of neuroimaging studies. The results have shown that emotional processing in depressed patients is topologically shifted towards the insular areas involved in pain perception and processing.26 At the molecular level, monoamine neurotransmitters, including serotonin, dopamine, and norepinephrine, have been found to typically involve in the pathologies of both depression and pain.15 For example, with depletion of serotonin and norepinephrine, as occurs in depression, the pain signals from the body, which are suppressed under normal conditions, are amplified with more attention and emotion involved.1 Moreover, common genetic and epigenetic modifications might also mediate the interaction between depression and pain. SNP Val66Met in the BNDF gene, which encodes for brain-derived neurotrophic factor, has been shown to modify the relationship between life stress and depression13 and increase the vulnerability to chronic multisite musculoskeletal pain.10 Rats experiencing stress-induced visceral pain have shown an increase in DNA methylation at the glucocorticoid receptor promoter and a decrease at the corticotropin-releasing factor genes in the amygdala, demonstrating the involvement of central epigenetic mechanisms in regulating pain-depression morbidity.31 Despite findings identified from animal models, our cell-type–specific annotation analysis, which leveraged human genome-wide information, has demonstrated a substantial enrichment in brain tissues of depression, headache, and neck/shoulder pain, supporting the hypothesis of neurobiological mechanisms underlying depression and pain.
Our current study has several advantages. We controlled for bias arising from population stratification by restricting participants to individuals of European ancestry. We conducted several important sensitivity analyses to verify MR model assumptions. We selected the most significant independent SNPs identified by the largest depression or pain GWAS, so all were robustly associated with exposure of interest, guaranteeing “relevance” assumption. We performed several statistical approaches to reduce pleiotropic effects and to satisfy “exclusion restriction” and “exchangeability” assumption.
Nevertheless, limitations should be acknowledged. First, despite using the hitherto largest GWAS for MDD and 7 localized pain, our statistical power remains limited for relatively small odds ratio detected in each association. In addition, overfitting might be a concern because both MDD and pain GWAS contain UK Biobank data (up to 28% participant overlap). We conducted a sensitivity analysis using 44 MDD-associated SNPs identified in an early GWAS possessing negligible overlap with pain GWAS and observed highly consistent results with our main analysis. Such consistency reinforced the robustness of our findings and reduced the likelihood of overfitting and false positives to the minimal. Furthermore, the pain phenotype in UK Biobank was based on a single uniformed question. This means that all pain phenotypes were broadly defined and self-reported, unfiltered by other potentially relevant information on the nature, duration, or intensity of the pain. Similar concerns apply to the depression measurement. An updated analysis is warranted when the new and more detailed, validated pain-related questionnaire will be administered. Also, the original pain GWAS(s) did not take into account whether participants were taking antidepressants when answering their pain questionnaires. Antidepressants have been found to relieve pain in depressed patients, which might misclassify pain cases into nonpain controls. However, the impact of such misclassification might be small due to 2 reasons. First, evidence from randomized clinical trials has demonstrated that antidepressants can relieve but can hardly eliminate pain, meaning individuals under treatment could still experience moderate pain feelings.12,18 Given the pain phenotypes in the original GWAS(s) were defined as binary questions (having vs not having pain), most of the participants with antidepressant treatment could still report a “Yes” in their pain questionnaires and contribute correctly as a case to the GWAS(s). Second, such misclassification, if exists, would drive our pain-MDD relationship towards null, contrary to the significant results observed by our study. Moreover, due to the minimal significant genetic signals that have been identified contributing to the pain at body sites except headache, our analysis targeting the pain-on-depression relationship was limited to headache only. An updated analysis is necessitated for the effects of site-specific pain on depression when more signals are discovered. Finally, because the participants in our present study were of European ancestry, we cautioned that the generalizability of our results is confined to European-ancestral populations.
To conclude, we have identified a causal connection between depression and pain at specific body sites such as head, neck/shoulder, back, and abdominal/stomach, but not at other body sites including face, hip, or knee. We have further revealed a brain tissue enrichment in depression, headache, and neck/should pain, which suggests possible common neurological pathways underpinning the causal association. Our findings contribute to the understanding of genetic and biological mechanisms for individual pain phenotypes and depression.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B367.
Supplementary Material
Acknowledgments
Summary statistics for the genetic associations with MDD, seven localized pain, multisite chronic pain, and five lifestyle factors were obtained from the corresponding GWAS listed in Supplementary Table 4 (available at http://links.lww.com/PAIN/B367). The authors thank all the investigators for sharing the data. XJ designed the study. BT and XJ analyzed the data, interpreted the results, and wrote the manuscript. Dr. Xia Jiang is supported by a starting package from the Swedish Research Council and Forskningsrådet för hälsa, arbetsliv och välfärd.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painjournalonline.com).
B. Tang and W. Meng contributed equally.
References
- [1].Bair MJ, Robinson RL, Katon W, Kroenke K. Depression and pain comorbidity: a literature review. Arch Intern Med 2003;163:2433–45. [DOI] [PubMed] [Google Scholar]
- [2].Blaauw BA, Dyb G, Hagen K, Holmen TL, Linde M, Wentzel-Larsen T, Zwart JA. The relationship of anxiety, depression and behavioral problems with recurrent headache in late adolescence—a Young-HUNT follow-up study. The J Headache Pain 2015;16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM. Schizophrenia Working Group of the Psychiatric Genomics C. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol 2015;181:251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Doherty A, Smith-Byrne K, Ferreira T, Holmes MV, Holmes C, Pulit SL, Lindgren CM. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun 2018;9:5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, Ripke S, Day FR, Purcell S, Stahl E, Lindstrom S, Perry JRB, Okada Y, Raychaudhuri S, Daly MJ, Patterson N, Neale BM, Price AL, ReproGen C. Schizophrenia Working Group of the Psychiatric Genomics C, the RC. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015;47:1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Generaal E, Milaneschi Y, Jansen R, Elzinga BM, Dekker J, Penninx BW. The brain-derived neurotrophic factor pathway, life stress, and chronic multi-site musculoskeletal pain. Mol Pain 2016;12:1744806916646783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Holroyd KA, O'Donnell FJ, Stensland M, Lipchik GL, Cordingley GE, Carlson BW. Management of chronic tension-type headache with tricyclic antidepressant medication, stress management therapy, and their combination: a randomized controlled trial. JAMA 2001;285:2208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hosang GM, Shiles C, Tansey KE, McGuffin P, Uher R. Interaction between stress and the BDNFVal66Met polymorphism in depression: a systematic review and meta-analysis. BMC Med 2014;12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, Alloza C, Shen X, Barbu MC, Xu EY, Whalley HC, Marioni RE, Porteous DJ, Davies G, Deary IJ, Hemani G, Berger K, Teismann H, Rawal R, Arolt V, Baune BT, Dannlowski U, Domschke K, Tian C, Hinds DA, Trzaskowski M, Byrne EM, Ripke S, Smith DJ, Sullivan PF, Wray NR, Breen G, Lewis CM, McIntosh AM, andMe Research T. Major Depressive Disorder Working Group of the Psychiatric Genomics C. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 2019;22:343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Humo M, Lu H, Yalcin I. The molecular neurobiology of chronic pain-induced depression. Cell Tissue Res 2019;377:21–43. [DOI] [PubMed] [Google Scholar]
- [16].Johnston KJA, Adams MJ, Nicholl BI, Ward J, Strawbridge RJ, Ferguson A, McIntosh AM, Bailey MES, Smith DJ. Genome-wide association study of multisite chronic pain in UK Biobank. Plos Genet 2019;15:e1008164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kindler S, Samietz S, Houshmand M, Grabe HJ, Bernhardt O, Biffar R, Kocher T, Meyer G, Völzke H, Metelmann H-R, Schwahn C. Depressive and anxiety symptoms as risk factors for temporomandibular joint pain: a prospective cohort study in the general population. J Pain 2012;13:1188–97. [DOI] [PubMed] [Google Scholar]
- [18].Kroenke K, Bair MJ, Damush TM, Wu J, Hoke S, Sutherland J, Tu W. Optimized antidepressant therapy and pain self-management in primary care patients with depression and musculoskeletal pain: a randomized controlled trial. JAMA 2009;301:2099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linnér R, Fontana MA, Kundu T, Lee C, Li H, Li R, Royer R, Timshel PN, Walters RK, Willoughby EA, Yengo L, andMe Research T, CogentSocial Science Genetic Association C, Alver M, Bao Y, Clark DW, Day FR, Furlotte NA, Joshi PK, Kemper KE, Kleinman A, Langenberg C, Mägi R, Trampush JW, Verma SS, Wu Y, Lam M, Zhao JH, Zheng Z, Boardman JD, Campbell H, Freese J, Harris KM, Hayward C, Herd P, Kumari M, Lencz T, Luan Ja, Malhotra AK, Metspalu A, Milani L, Ong KK, Perry JRB, Porteous DJ, Ritchie MD, Smart MC, Smith BH, Tung JY, Wareham NJ, Wilson JF, Beauchamp JP, Conley DC, Esko T, Lehrer SF, Magnusson PKE, Oskarsson S, Pers TH, Robinson MR, Thom K, Watson C, Chabris CF, Meyer MN, Laibson DI, Yang J, Johannesson M, Koellinger PD, Turley P, Visscher PM, Benjamin DJ, Cesarini D. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 2018;50:1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li JX. Pain and depression comorbidity: a preclinical perspective. Behav Brain Res 2015;276:92–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hinds DA, Hromatka BS, Huber KE, Kleinman A, Litterman NK, McIntyre MH, Mountain JL, Northover CAM, Sathirapongsasuti JF, Sazonova OV, Shelton JF, Shringarpure S, Tian C, Tung JY, Vacic V, Wilson CH, Pitts SJ, Mitchell A, Skogholt AH, Winsvold BS, Sivertsen B, Stordal E, Morken G, Kallestad H, Heuch I, Zwart JA, Fjukstad KK, Pedersen LM, Gabrielsen ME, Johnsen MB, Skrove M, Indredavik MS, Drange OK, Bjerkeset O, Børte S, Stensland SØ, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga J-J, Huang H, Jang SK, Jansen PR, Ling Y, Mägi R, Matoba N, McMahon G, Mulas A, Orrù V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stančáková A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafò MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S, andMe Research T, Psychiatry HA-I. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019;51:237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Meng W, Adams MJ, Hebert HL, Deary IJ, McIntosh AM, Smith BH. A genome-wide association study finds genetic associations with broadly-defined headache in UK Biobank (N=223,773). EBioMedicine 2018;28:180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Meng W, Adams MJ, Palmer CNA, Agee M, Alipanahi B, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hicks B, Hinds DA, Huber KE, Jewett EM, Jiang Y, Kleinman A, Lin K-H, Litterman NK, McCreight JC, McIntyre MH, McManus KF, Mountain JL, Noblin ES, Northover CAM, Pitts SJ, Poznik GD, Sathirapongsasuti JF, Shelton JF, Shringarpure S, Tian C, Tung JY, Vacic V, Wang X, Wilson CH, Shi J, Auton A, Ryan KA, Jordan JM, Mitchell BD, Jackson RD, Yau MS, McIntosh AM, Smith BH. The 23andMe Research T. Genome-wide association study of knee pain identifies associations with GDF5 and COL27A1 in UK Biobank. Commun Biol 2019;2:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Meng W, Adams MJ, Reel P, Rajendrakumar A, Huang Y, Deary IJ, Palmer CNA, McIntosh AM, Smith BH. Genetic correlations between pain phenotypes and depression and neuroticism. Eur J Hum Genet 2020;28:358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Meng W, Chan BW, Harris C, Freidin MB, Hebert HL, Adams MJ, Campbell A, Hayward C, Zheng H, Zhang X, Colvin LA, Hales TG, Palmer CNA, Williams FMK, McIntosh A, Smith BH. A genome-wide association study finds genetic variants associated with neck or shoulder pain in UK Biobank. Hum Mol Genet 2020;29:1396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mutschler I, Ball T, Wankerl J, Strigo IA. Pain and emotion in the insular cortex: evidence for functional reorganization in major depression. Neurosci Lett 2012;520:204–9. [DOI] [PubMed] [Google Scholar]
- [27].Riddle DL, Kong X, Fitzgerald GK. Psychological health impact on 2-year changes in pain and function in persons with knee pain: data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2011;19:1095–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rist PM, Schürks M, Buring JE, Kurth T. Migraine, headache, and the risk of depression: prospective cohort study. Cephalalgia 2013;33:1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schwarze M, Häuser W, Schmutzer G, Brähler E, Beckmann NA, Schiltenwolf M. Obesity, depression and hip pain. Musculoskeletal Care 2019;17:126–32. [DOI] [PubMed] [Google Scholar]
- [30].Suri P, Palmer MR, Tsepilov YA, Freidin MB, Boer CG, Yau MS, Evans DS, Gelemanovic A, Bartz TM, Nethander M, Arbeeva L, Karssen L, Neogi T, Campbell A, Mellstrom D, Ohlsson C, Marshall LM, Orwoll E, Uitterlinden A, Rotter JI, Lauc G, Psaty BM, Karlsson MK, Lane NE, Jarvik GP, Polasek O, Hochberg M, Jordan JM, Van Meurs JBJ, Jackson R, Nielson CM, Mitchell BD, Smith BH, Hayward C, Smith NL, Aulchenko YS, Williams FMK. Genome-wide meta-analysis of 158,000 individuals of European ancestry identifies three loci associated with chronic back pain. Plos Genet 2018;14:e1007601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tran L, Chaloner A, Sawalha AH, Greenwood Van-Meerveld B. Importance of epigenetic mechanisms in visceral pain induced by chronic water avoidance stress. Psychoneuroendocrinology 2013;38:898–906. [DOI] [PubMed] [Google Scholar]
- [32].Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, Bacanu S-A, Bækvad-Hansen M, Beekman AFT, Bigdeli TB, Binder EB, Blackwood DRH, Bryois J, Buttenschøn HN, Bybjerg-Grauholm J, Cai N, Castelao E, Christensen JH, Clarke T-K, Coleman JIR, Colodro-Conde L, Couvy-Duchesne B, Craddock N, Crawford GE, Crowley CA, Dashti HS, Davies G, Deary IJ, Degenhardt F, Derks EM, Direk N, Dolan CV, Dunn EC, Eley TC, Eriksson N, Escott-Price V, Kiadeh FHF, Finucane HK, Forstner AJ, Frank J, Gaspar HA, Gill M, Giusti-Rodríguez P, Goes FS, Gordon SD, Grove J, Hall LS, Hannon E, Hansen CS, Hansen TF, Herms S, Hickie IB, Hoffmann P, Homuth G, Horn C, Hottenga J-J, Hougaard DM, Hu M, Hyde CL, Ising M, Jansen R, Jin F, Jorgenson E, Knowles JA, Kohane IS, Kraft J, Kretzschmar WW, Krogh J, Kutalik Z, Lane JM, Li Y, Li Y, Lind PA, Liu X, Lu L, MacIntyre DJ, MacKinnon DF, Maier RM, Maier W, Marchini J, Mbarek H, McGrath P, McGuffin P, Medland SE, Mehta D, Middeldorp CM, Mihailov E, Milaneschi Y, Milani L, Mill J, Mondimore FM, Montgomery GW, Mostafavi S, Mullins N, Nauck M, Ng B. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018;50:668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B367.


