Extended Data Fig. 8 |. IDH-MUT cells exhibit preferential enhancer hypermethylation, decoupling of the promoter methylation-expression relationship and disruption of CTCF insulation.
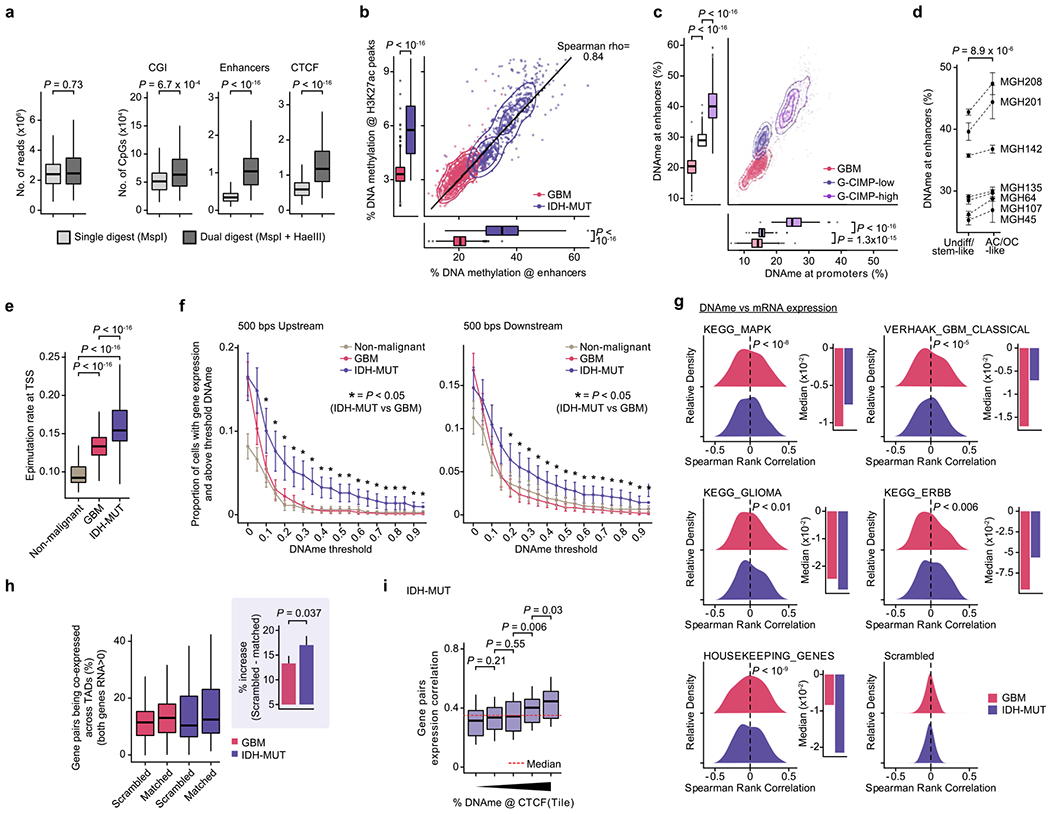
a, Number of aligned reads and unique CpGs for MspI (n=476) and HaeIII+MspI digested IDH-MUT cells (n=242; MGH201 and MGH208). b, Mean CpG methylation at FANTOM5 enhancers vs. H3K27ac ChIP-seq peaks47,70 between GBM (n=765) and IDH-MUT (n=670) cells. c, Mean CpG methylation at TSS (±1Kb) vs. FANTOM5 enhancers between GBM (n=765) and IDH-MUT (n=670) cells (G-CIMP-low [MGH107, MGH135, MGH45, MGH64]; G-CIMP-high [MGH142, MGH201, MGH208]). d, Mean (±SEM) CpG methylation at FANTOM5 enhancers for stem-like/undifferentiated and AC-/OC-like IDH-MUT cells. e, Epimutation rate across non-malignant (n=148), GBM (n=765) and IDH-MUT (n=670) cells. f, Proportion of cells with gene expression (read count >0) and above-threshold DNAme at 500 base-pairs regions upstream (left) or downstream (right) of TSS. Data are mean (±s.e.m.) across all genes (expression seen in > 5 cells, DNAme >5 CpGs per region) for non-malignant cells (n=148), GBM (n=765) and IDH-MUT (n=670) cells. ’*’ P-value < 0.05. g, Left: Distribution of Spearman’s rho of expression and promoter DNAme correlation (n=1,523 genes expressed >5 cells, DNAme >5 CpGs per promoter); GBM (n=765) and IDH-MUT (n=670) cells. Right: Median values of Spearman’s rho of expression and promoter DNAme correlation. h, Percentage of genes pairs across CTCF sites70 being co-expressed (both RNA read count >0); GBM (n=765) and IDH-MUT (n=670) cells. Scrambled represents randomly permuted cell labels for the expression values. Inset: Increase in percentage of genes pairs across CTCF sites70 being co-expressed when comparing matched vs. scrambled groups. Error bars represent 95% CIs. i, Gene expression correlation (Spearman’s rho) of genes pairs across CTCF sites70 per tile of mean CpG methylation at CTCF binding sites (low-to-high); IDH-MUT (n=670) cells. P values are two-sided Mann-Whitney U test (a-c, e-f, h-i), Fisher’s combined probability test (d), two-sided Kolmogorov-Smirnov test (g). Boxplots represent the median, bottom and upper quartiles, whiskers correspond to 1.5 times the interquartile range.
